[English] 日本語
 Yorodumi
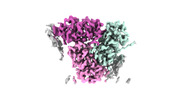
Yorodumi- EMDB-33737: Cryo-EM structure of the PSI-LHCI-Lhcp supercomplex from Ostreoco... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the PSI-LHCI-Lhcp supercomplex from Ostreococcus tauri | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
| Function / homology |  Function and homology information Function and homology information chloroplast photosystem I / chloroplast photosystem I /  photosynthesis, light harvesting in photosystem I / chloroplast thylakoid lumen / photosynthesis, light harvesting in photosystem I / chloroplast thylakoid lumen /  photosynthesis, light harvesting / photosynthesis, light harvesting /  photosystem I reaction center / photosystem I reaction center /  photosystem I / photosynthetic electron transport in photosystem I / photosystem I / photosynthetic electron transport in photosystem I /  photosystem I / photosystem I /  photosystem II / photosystem II /  chlorophyll binding ... chlorophyll binding ... chloroplast photosystem I / chloroplast photosystem I /  photosynthesis, light harvesting in photosystem I / chloroplast thylakoid lumen / photosynthesis, light harvesting in photosystem I / chloroplast thylakoid lumen /  photosynthesis, light harvesting / photosynthesis, light harvesting /  photosystem I reaction center / photosystem I reaction center /  photosystem I / photosynthetic electron transport in photosystem I / photosystem I / photosynthetic electron transport in photosystem I /  photosystem I / photosystem I /  photosystem II / photosystem II /  chlorophyll binding / chloroplast thylakoid membrane / response to light stimulus / chlorophyll binding / chloroplast thylakoid membrane / response to light stimulus /  photosynthesis / photosynthesis /  chloroplast / 4 iron, 4 sulfur cluster binding / chloroplast / 4 iron, 4 sulfur cluster binding /  electron transfer activity / magnesium ion binding / electron transfer activity / magnesium ion binding /  membrane / membrane /  metal ion binding metal ion bindingSimilarity search - Function | |||||||||||||||
| Biological species |   Ostreococcus tauri (plant) Ostreococcus tauri (plant) | |||||||||||||||
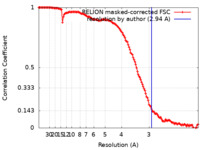
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.94 Å cryo EM / Resolution: 2.94 Å | |||||||||||||||
 Authors Authors | Shan J / Sheng X / Ishii A / Watanabe A / Song C / Murata K / Minagawa J / Liu Z | |||||||||||||||
| Funding support |  Japan, Japan,  China, 4 items China, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2023 Journal: Elife / Year: 2023Title: The photosystem I supercomplex from a primordial green alga harbors three light-harvesting complex trimers. Authors: Asako Ishii / Jianyu Shan / Xin Sheng / Eunchul Kim / Akimasa Watanabe / Makio Yokono / Chiyo Noda / Chihong Song / Kazuyoshi Murata / Zhenfeng Liu / Jun Minagawa /   Abstract: As a ubiquitous picophytoplankton in the ocean and an early-branching green alga, is a model prasinophyte species for studying the functional evolution of the light-harvesting systems in ...As a ubiquitous picophytoplankton in the ocean and an early-branching green alga, is a model prasinophyte species for studying the functional evolution of the light-harvesting systems in photosynthesis. Here, we report the structure and function of the photosystem I (PSI) supercomplex in low light conditions, where it expands its photon-absorbing capacity by assembling with the light-harvesting complexes I (LHCI) and a prasinophyte-specific light-harvesting complex (Lhcp). The architecture of the supercomplex exhibits hybrid features of the plant-type and the green algal-type PSI supercomplexes, consisting of a PSI core, an Lhca1-Lhca4-Lhca2-Lhca3 belt attached on one side and an Lhca5-Lhca6 heterodimer associated on the other side between PsaG and PsaH. Interestingly, nine Lhcp subunits, including one Lhcp1 monomer with a phosphorylated amino-terminal threonine and eight Lhcp2 monomers, oligomerize into three trimers and associate with PSI on the third side between Lhca6 and PsaK. The Lhcp1 phosphorylation and the light-harvesting capacity of PSI were subjected to reversible photoacclimation, suggesting that the formation of PSI-LHCI-Lhcp supercomplex is likely due to a phosphorylation-dependent mechanism induced by changes in light intensity. Notably, this supercomplex did not exhibit far-red peaks in the 77 K fluorescence spectra, which is possibly due to the weak coupling of the chlorophyll 603-609 pair in Lhca1-4. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33737.map.gz emd_33737.map.gz | 202.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33737-v30.xml emd-33737-v30.xml emd-33737.xml emd-33737.xml | 48.3 KB 48.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33737_fsc.xml emd_33737_fsc.xml | 13.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_33737.png emd_33737.png | 179 KB | ||
| Others |  emd_33737_half_map_1.map.gz emd_33737_half_map_1.map.gz emd_33737_half_map_2.map.gz emd_33737_half_map_2.map.gz | 171.4 MB 171.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33737 http://ftp.pdbj.org/pub/emdb/structures/EMD-33737 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33737 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33737 | HTTPS FTP |
-Related structure data
| Related structure data |  7ycaMC  8hg3C  8hg5C  8hg6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33737.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33737.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33737_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
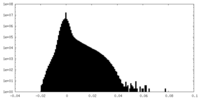
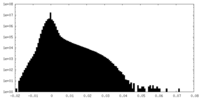
| Density Histograms |
-Half map: #1
| File | emd_33737_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
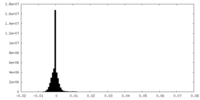
| Density Histograms |
- Sample components
Sample components
+Entire : photosystem I-light-harvesting complex I-prasinophyte-specific Lh...
+Supramolecule #1: photosystem I-light-harvesting complex I-prasinophyte-specific Lh...
+Macromolecule #1: Lhca1
+Macromolecule #2: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #3: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #4: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #5: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #6: Lhca6
+Macromolecule #7: Photosystem I P700 chlorophyll a apoprotein A1
+Macromolecule #8: Photosystem I P700 chlorophyll a apoprotein A2
+Macromolecule #9: Photosystem I iron-sulfur center
+Macromolecule #10: Photosystem I reaction center subunit II, chloroplastic
+Macromolecule #11: Photosystem I reaction centre subunit IV
+Macromolecule #12: Photosystem I reaction center subunit III
+Macromolecule #13: PsaG
+Macromolecule #14: Photosystem I PsaH, reaction centre subunit VI
+Macromolecule #15: Photosystem I reaction center subunit VIII
+Macromolecule #16: Photosystem I reaction center subunit IX
+Macromolecule #17: Photosystem I PsaG/PsaK protein
+Macromolecule #18: PSI subunit V
+Macromolecule #19: Photosystem I reaction center subunit XII
+Macromolecule #20: Photosystem I PsaN, reaction centre subunit N
+Macromolecule #21: PsaO
+Macromolecule #22: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #23: Chlorophyll a-b binding protein, chloroplastic
+Macromolecule #24: CHLOROPHYLL B
+Macromolecule #25: CHLOROPHYLL A
+Macromolecule #26: (3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-1-[(1~{S},4...
+Macromolecule #27: (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BE...
+Macromolecule #28: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE
+Macromolecule #29: (1~{S})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E}...
+Macromolecule #30: BETA-CAROTENE
+Macromolecule #31: 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE
+Macromolecule #32: 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL
+Macromolecule #33: CHLOROPHYLL A ISOMER
+Macromolecule #34: PHYLLOQUINONE
+Macromolecule #35: IRON/SULFUR CLUSTER
+Macromolecule #36: DIGALACTOSYL DIACYL GLYCEROL (DGDG)
+Macromolecule #37: (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY...
+Macromolecule #38: Chlorophyll c2
+Macromolecule #39: water
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 |
| Vitrification | Cryogen name: NITROGEN / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.8 µm Bright-field microscopy / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.8 µm |
| Image recording | Film or detector model: GATAN K2 BASE (4k x 4k) / Average electron dose: 60.0 e/Å2 |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | (Chain: PDB, experimental model, PDB, experimental model, PDB, experimental model, PDB, experimental model, PDB, experimental model, PDB, experimental model, PDB, experimental model, PDB, experimental model, PDB, experimental model, PDB, experimental model, PDB, experimental model, PDB, experimental model, PDB, experimental model, PDB, experimental model, PDB, experimental model) |
|---|---|
| Details | Initial local fitting was done using Chimera and then Wincoot was usd for flexible fitting |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 110.967 / Target criteria: Correlation coefficient |
| Output model |  PDB-7yca: |
 Movie
Movie Controller
Controller














 Z
Z Y
Y X
X