[English] 日本語
 Yorodumi
Yorodumi- EMDB-29978: McrD binds asymmetrically to methyl-coenzyme M reductase improvin... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 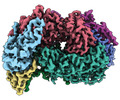 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
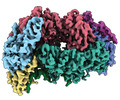
| Title | McrD binds asymmetrically to methyl-coenzyme M reductase improving active site accessibility during assembly | ||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords |  methanogenesis / MCR complex / methanogenesis / MCR complex /  TRANSFERASE TRANSFERASE | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology information coenzyme-B sulfoethylthiotransferase / coenzyme-B sulfoethylthiotransferase /  coenzyme-B sulfoethylthiotransferase activity / coenzyme-B sulfoethylthiotransferase activity /  methanogenesis / methanogenesis /  metal ion binding / metal ion binding /  cytoplasm cytoplasmSimilarity search - Function | ||||||||||||||||||||||||
| Biological species |   Methanosarcina acetivorans C2A (archaea) Methanosarcina acetivorans C2A (archaea) | ||||||||||||||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.0 Å cryo EM / Resolution: 3.0 Å | ||||||||||||||||||||||||
 Authors Authors | Joiner AMN / Chadwick GL / Nayak DD | ||||||||||||||||||||||||
| Funding support |  United States, 7 items United States, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: McrD binds asymmetrically to methyl-coenzyme M reductase improving active-site accessibility during assembly. Authors: Grayson L Chadwick / Aaron M N Joiner / Sangeetha Ramesh / Douglas A Mitchell / Dipti D Nayak /  Abstract: Methyl-coenzyme M reductase (MCR) catalyzes the formation of methane, and its activity accounts for nearly all biologically produced methane released into the atmosphere. The assembly of MCR is an ...Methyl-coenzyme M reductase (MCR) catalyzes the formation of methane, and its activity accounts for nearly all biologically produced methane released into the atmosphere. The assembly of MCR is an intricate process involving the installation of a complex set of posttranslational modifications and the unique Ni-containing tetrapyrrole called coenzyme F. Despite decades of research, details of MCR assembly remain largely unresolved. Here, we report the structural characterization of MCR in two intermediate states of assembly. These intermediate states lack one or both F cofactors and form complexes with the previously uncharacterized McrD protein. McrD is found to bind asymmetrically to MCR, displacing large regions of the alpha subunit and increasing active-site accessibility for the installation of F-shedding light on the assembly of MCR and the role of McrD therein. This work offers crucial information for the expression of MCR in a heterologous host and provides targets for the design of MCR inhibitors. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29978.map.gz emd_29978.map.gz | 21.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29978-v30.xml emd-29978-v30.xml emd-29978.xml emd-29978.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29978_fsc.xml emd_29978_fsc.xml | 7.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_29978.png emd_29978.png | 171.6 KB | ||
| Others |  emd_29978_half_map_1.map.gz emd_29978_half_map_1.map.gz emd_29978_half_map_2.map.gz emd_29978_half_map_2.map.gz | 39.8 MB 39.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29978 http://ftp.pdbj.org/pub/emdb/structures/EMD-29978 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29978 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29978 | HTTPS FTP |
-Related structure data
| Related structure data |  8gf5MC  8gf6C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29978.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29978.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.115 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_29978_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_29978_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Assembly intermediate of the MCR complex bound to mcrD
| Entire | Name: Assembly intermediate of the MCR complex bound to mcrD |
|---|---|
| Components |
|
-Supramolecule #1: Assembly intermediate of the MCR complex bound to mcrD
| Supramolecule | Name: Assembly intermediate of the MCR complex bound to mcrD type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Methanosarcina acetivorans C2A (archaea) Methanosarcina acetivorans C2A (archaea) |
-Macromolecule #1: Methyl-coenzyme M reductase subunit alpha
| Macromolecule | Name: Methyl-coenzyme M reductase subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number:  coenzyme-B sulfoethylthiotransferase coenzyme-B sulfoethylthiotransferase |
|---|---|
| Source (natural) | Organism:   Methanosarcina acetivorans C2A (archaea) Methanosarcina acetivorans C2A (archaea) |
| Molecular weight | Theoretical: 62.180078 KDa |
| Sequence | String: MAADIFAKFK KSMEVKFTQE YGSNKQAGGD ITGKTEKFLR LGPEQDARKQ EMIKAGKEIA EKRGIAFYNP MMHMGAPLGQ RAITPYTIS GTDIVAEPDD LHYVNNAAMQ QMWDDIRRTC IVGLDMAHET LEKRLGKEVT PETINHYLET LNHAMPGAAV V QEMMVETH ...String: MAADIFAKFK KSMEVKFTQE YGSNKQAGGD ITGKTEKFLR LGPEQDARKQ EMIKAGKEIA EKRGIAFYNP MMHMGAPLGQ RAITPYTIS GTDIVAEPDD LHYVNNAAMQ QMWDDIRRTC IVGLDMAHET LEKRLGKEVT PETINHYLET LNHAMPGAAV V QEMMVETH PALVDDCYVK IFTGDDELAD EIDKQYVINV NKMFSEEQAA QIKASIGKTT WQAIHIPTIV SRTTDGAQTS RW AAMQIGM SFISAYAMCA GEAAVADLSF AAK(MHS)AALVSM GEMLPAR(AGM)AR GPNEPGGLSF GHLSDIVQTS RVSKD PAKI ALEVVGAGCM LYDQIWLGSY MSGGVGFTQY ATAAYTDDIL DNNTYYDVDY INDKYNGAAN LGTDNKVKAT LDVVKD IAT ESTLYGIETY EKFPTALEDH FGGSQRATVL AAASGVACAL ATGNANAGLS GWYLSMYVHK EAWGRLGFFG FDLQDQ (SMC)GA TNVLSYQGDE GLPDELRGPN YPNYAMNVGH QGGYAGIAQA AHSGRGDAFT VNPLLKVCFA DELMPFNFAE PRR EFGRGA IREFMPAGER SLVIPAK |
-Macromolecule #2: Methyl-coenzyme M reductase subunit beta
| Macromolecule | Name: Methyl-coenzyme M reductase subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Methanosarcina acetivorans C2A (archaea) Methanosarcina acetivorans C2A (archaea) |
| Molecular weight | Theoretical: 45.17407 KDa |
| Sequence | String: MSDTVDIYDD RGKLLESNVD IMSLAPTRNA AIKKIILDTK RSVAVSLAGI QGALASGKMG GKGRQILGRG LNYDLVGNAD AIAENVKNL VQVDEGDDTS VKVIKGGKSL LIQAPSSRIA AGADYMSATT VGAAAVTQTI IDMFGTDMYD APIAKSAVWG S YPQTMDLM ...String: MSDTVDIYDD RGKLLESNVD IMSLAPTRNA AIKKIILDTK RSVAVSLAGI QGALASGKMG GKGRQILGRG LNYDLVGNAD AIAENVKNL VQVDEGDDTS VKVIKGGKSL LIQAPSSRIA AGADYMSATT VGAAAVTQTI IDMFGTDMYD APIAKSAVWG S YPQTMDLM GGNVQGVLSI PQNNEGLGFS LRNIMANHIA AITSRGAMNA AALSSIYEQS GIFEMGGAVG MFERHQLLGL AC QGLNANN VVYDIVKENG KDGTIGTVIE SIVGRAVEDG VISVDKTAPS GYKFYKANDV PMWNAYAAAG TLAATFVNCG AGR AAQNVS STLLYFNDIL EKETGLPGCD YGKVQGVAVG FSFFSHSIYG GGGPGVFNGN HVVTRHSRGF AIPCVCAAVA LDAG TQMFT IESTSGLIGD VFGSIEEFRQ PIKAVAGAL |
-Macromolecule #3: Methyl-coenzyme M reductase subunit gamma
| Macromolecule | Name: Methyl-coenzyme M reductase subunit gamma / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Methanosarcina acetivorans C2A (archaea) Methanosarcina acetivorans C2A (archaea) |
| Molecular weight | Theoretical: 27.630184 KDa |
| Sequence | String: MAYEAQYYPG ATSVGANRRK HMSGKLEKLR EISDEDLTAV LGHRAPGSDY PSTHPPLAEM GEPACSIREA VAATPGAAAG DRVRYVQFA DSMYNAPATP YFRSYFAAIN FRGVDPGTLS GRQIVEARER DMEQCAKVQM ETEMTDPALA GMRGATVHGH S VRLQEDGV ...String: MAYEAQYYPG ATSVGANRRK HMSGKLEKLR EISDEDLTAV LGHRAPGSDY PSTHPPLAEM GEPACSIREA VAATPGAAAG DRVRYVQFA DSMYNAPATP YFRSYFAAIN FRGVDPGTLS GRQIVEARER DMEQCAKVQM ETEMTDPALA GMRGATVHGH S VRLQEDGV MFDMLDRRRL EGGVIIMDKD QVAIPLDRKV NLGKPMSSEE AAKRTTIYRV DNVAFRDDAE VIEWVHRVFD QR TSYGFQP K |
-Macromolecule #4: Methyl coenzyme M reductase, subunit D
| Macromolecule | Name: Methyl coenzyme M reductase, subunit D / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Methanosarcina acetivorans C2A (archaea) Methanosarcina acetivorans C2A (archaea) |
| Molecular weight | Theoretical: 21.842742 KDa |
| Recombinant expression | Organism:   Methanosarcina acetivorans C2A (archaea) Methanosarcina acetivorans C2A (archaea) |
| Sequence | String: MDYKDDDDKG GGWSHPQFEK GGGMSDSASN TEDSIQIEIF PSRILSPETA QKLISELYQV DGIIRVMVQG PRLPERVSAG PGTGEKVEH PLRKPIQIGD QVIELKISVG RIRLEIENAE TKEKVRSVCD KMLPFSFEFR EGHFLRRKPT VTDYAKLGPE T DPRLLGMV ...String: MDYKDDDDKG GGWSHPQFEK GGGMSDSASN TEDSIQIEIF PSRILSPETA QKLISELYQV DGIIRVMVQG PRLPERVSAG PGTGEKVEH PLRKPIQIGD QVIELKISVG RIRLEIENAE TKEKVRSVCD KMLPFSFEFR EGHFLRRKPT VTDYAKLGPE T DPRLLGMV DPKAKVNQLV FIEKREKEDD TDKDE |
-Macromolecule #5: 1-THIOETHANESULFONIC ACID
| Macromolecule | Name: 1-THIOETHANESULFONIC ACID / type: ligand / ID: 5 / Number of copies: 1 / Formula: COM |
|---|---|
| Molecular weight | Theoretical: 142.197 Da |
| Chemical component information |  ChemComp-COM: |
-Macromolecule #6: FACTOR 430
| Macromolecule | Name: FACTOR 430 / type: ligand / ID: 6 / Number of copies: 1 / Formula: F43 |
|---|---|
| Molecular weight | Theoretical: 906.58 Da |
| Chemical component information | 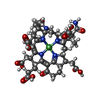 ChemComp-F43: |
-Macromolecule #7: Coenzyme B
| Macromolecule | Name: Coenzyme B / type: ligand / ID: 7 / Number of copies: 1 / Formula: TP7 |
|---|---|
| Molecular weight | Theoretical: 343.334 Da |
| Chemical component information |  ChemComp-TP7: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.55 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: Vitrification occurred under aerobic conditions. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 36000 Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 36000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z
Z Y
Y X
X


















