+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SIV E660.CR54 SOS-2P Env Trimer with ITS92.02 | |||||||||
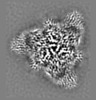
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationmembrane fusion involved in viral entry into host cell / host cell endosome membrane / membrane => GO:0016020 / symbiont entry into host cell /  viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |    Influenza A virus / Influenza A virus /   Simian immunodeficiency virus / Simian immunodeficiency virus /   Macaca mulatta (Rhesus monkey) Macaca mulatta (Rhesus monkey) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.32 Å cryo EM / Resolution: 4.32 Å | |||||||||
 Authors Authors | Gorman J / Kwong PD | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Cryo-EM structures of prefusion SIV envelope trimer. Authors: Jason Gorman / Chunyan Wang / Rosemarie D Mason / Alexandra F Nazzari / Hugh C Welles / Tongqing Zhou / Julian W Bess / Tatsiana Bylund / Myungjin Lee / Yaroslav Tsybovsky / Raffaello ...Authors: Jason Gorman / Chunyan Wang / Rosemarie D Mason / Alexandra F Nazzari / Hugh C Welles / Tongqing Zhou / Julian W Bess / Tatsiana Bylund / Myungjin Lee / Yaroslav Tsybovsky / Raffaello Verardi / Shuishu Wang / Yongping Yang / Baoshan Zhang / Reda Rawi / Brandon F Keele / Jeffrey D Lifson / Jun Liu / Mario Roederer / Peter D Kwong /  Abstract: Simian immunodeficiency viruses (SIVs) are lentiviruses that naturally infect non-human primates of African origin and seeded cross-species transmissions of HIV-1 and HIV-2. Here we report prefusion ...Simian immunodeficiency viruses (SIVs) are lentiviruses that naturally infect non-human primates of African origin and seeded cross-species transmissions of HIV-1 and HIV-2. Here we report prefusion stabilization and cryo-EM structures of soluble envelope (Env) trimers from rhesus macaque SIV (SIV) in complex with neutralizing antibodies. These structures provide residue-level definition for SIV-specific disulfide-bonded variable loops (V1 and V2), which we used to delineate variable-loop coverage of the Env trimer. The defined variable loops enabled us to investigate assembled Env-glycan shields throughout SIV, which we found to comprise both N- and O-linked glycans, the latter emanating from V1 inserts, which bound the O-link-specific lectin jacalin. We also investigated in situ SIV-Env trimers on virions, determining cryo-electron tomography structures at subnanometer resolutions for an antibody-bound complex and a ligand-free state. Collectively, these structures define the prefusion-closed structure of the SIV-Env trimer and delineate variable-loop and glycan-shielding mechanisms of immune evasion conserved throughout SIV evolution. | |||||||||
| History |
|
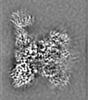
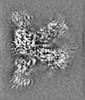
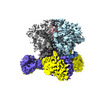
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27718.map.gz emd_27718.map.gz | 117.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27718-v30.xml emd-27718-v30.xml emd-27718.xml emd-27718.xml | 23.3 KB 23.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27718.png emd_27718.png | 88.6 KB | ||
| Masks |  emd_27718_msk_1.map emd_27718_msk_1.map | 125 MB |  Mask map Mask map | |
| Others |  emd_27718_additional_1.map.gz emd_27718_additional_1.map.gz emd_27718_additional_2.map.gz emd_27718_additional_2.map.gz emd_27718_half_map_1.map.gz emd_27718_half_map_1.map.gz emd_27718_half_map_2.map.gz emd_27718_half_map_2.map.gz | 17.8 MB 24.3 MB 116.2 MB 116.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27718 http://ftp.pdbj.org/pub/emdb/structures/EMD-27718 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27718 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27718 | HTTPS FTP |
-Related structure data
| Related structure data |  8duaMC  8dvdC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27718.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27718.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.083 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
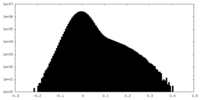
-Mask #1
| File |  emd_27718_msk_1.map emd_27718_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
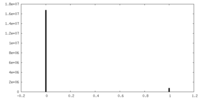
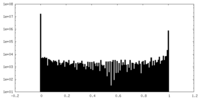
| Projections & Slices |
| ||||||||||||
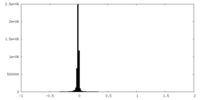
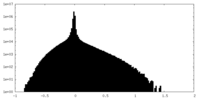
| Density Histograms |
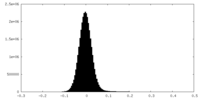
-Additional map: density modified map
| File | emd_27718_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | density modified map | ||||||||||||
| Projections & Slices |
| ||||||||||||
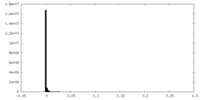
| Density Histograms |
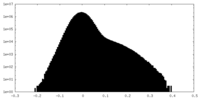
-Additional map: unsharpened map
| File | emd_27718_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
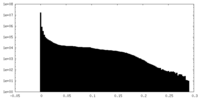
| Density Histograms |
-Half map: #2
| File | emd_27718_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
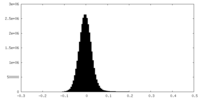
| Density Histograms |
-Half map: #1
| File | emd_27718_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SIV E660.CR54 SOS-2P with ITS92.02
| Entire | Name: SIV E660.CR54 SOS-2P with ITS92.02 |
|---|---|
| Components |
|
-Supramolecule #1: SIV E660.CR54 SOS-2P with ITS92.02
| Supramolecule | Name: SIV E660.CR54 SOS-2P with ITS92.02 / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:    Influenza A virus Influenza A virus |
-Macromolecule #1: Envelope glycoprotein gp41
| Macromolecule | Name: Envelope glycoprotein gp41 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Simian immunodeficiency virus Simian immunodeficiency virus |
| Molecular weight | Theoretical: 16.852207 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GVFVLGFLGF LATAGSAMGA ASLTLSAQSR TLLAGIVQQQ QQLLDVPKRQ QELLRLPVWG TKNLQTRVTA IEKYLKDQAQ LNSWGCAFR QVCCTTVPWP NETLVPNWSN MTWQEWERQV DFLEANITQL LEEAQIQQEK NMYELQKLN |
-Macromolecule #2: Envelope glycoprotein gp120
| Macromolecule | Name: Envelope glycoprotein gp120 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Simian immunodeficiency virus Simian immunodeficiency virus |
| Molecular weight | Theoretical: 59.053578 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: IQYVTVFYGV PAWKNATIPL FCATKNRDTW GTTQCLPDND DYSELAINIT EAFDAWNNTV TEQAIEDVWN LFETSIKPCV KLTPLCIAM RCNKTETDRW GLTRNAGTTT TTTTTTTAAT PSVAENVINE SNPCIKNNSC AGLEQEPMIG CKFNMTGLKR D KRIEYNET ...String: IQYVTVFYGV PAWKNATIPL FCATKNRDTW GTTQCLPDND DYSELAINIT EAFDAWNNTV TEQAIEDVWN LFETSIKPCV KLTPLCIAM RCNKTETDRW GLTRNAGTTT TTTTTTTAAT PSVAENVINE SNPCIKNNSC AGLEQEPMIG CKFNMTGLKR D KRIEYNET WYSRDLICEQ SANESESKCY MHHCNTSVIQ ESCDKHYWDA IRFRYCAPPG YALLRCNDSN YSGFAPNCSK VV VSSCTRM METQTSTWFG FNGTRAENRT YIYWHGKSNR TIISLNKYYN LTMRCRRPGN KTVLPVTIMS GLVFHSQPIN ERP KQAWCW FGGSWKEAIQ EVKETLVKHP RYTGTNDTRK INLTAPAGGD PEVTFMWTNC RGEFLYCKMN WFLNWVEDRD QKSS RWRQQ NTRERQKKNY VPCHIRQIIN TWHKVGKNVY LPPREGDLTC NSTVTSLIAE IDWTNNNETN ITMSAEVAEL YRLEL GDYK LVEITPIGLA PTSCRRYTTT GASRRRRR |
-Macromolecule #3: ITS92.02 Heavy Chain
| Macromolecule | Name: ITS92.02 Heavy Chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Macaca mulatta (Rhesus monkey) Macaca mulatta (Rhesus monkey) |
| Molecular weight | Theoretical: 24.275268 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLQESGPG LVKPSETLSL TCAVSGGSVS GNWWSWIRQP PGKGLEYIGR ISGSDGTTYY RSSLKVTISR DTSKNQFSLR LNSVTAADT AVYYCARKPT WDWFDVWGPG LLVTVSSAST KGPSVFPLAP SSKSTSGGTA ALGCLVKDYF PEPVTVSWNS G ALTSGVHT ...String: QVQLQESGPG LVKPSETLSL TCAVSGGSVS GNWWSWIRQP PGKGLEYIGR ISGSDGTTYY RSSLKVTISR DTSKNQFSLR LNSVTAADT AVYYCARKPT WDWFDVWGPG LLVTVSSAST KGPSVFPLAP SSKSTSGGTA ALGCLVKDYF PEPVTVSWNS G ALTSGVHT FPAVLQSSGL YSLSSVVTVP SSSLGTQTYI CNVNHKPSNT KVDKKVEPKS CDKGLEVLFQ |
-Macromolecule #4: ITS92.02 Light Chain
| Macromolecule | Name: ITS92.02 Light Chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Macaca mulatta (Rhesus monkey) Macaca mulatta (Rhesus monkey) |
| Molecular weight | Theoretical: 23.397963 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS LSASVGDRVT ITCRASQGID NYLNWYQQKP GKAPKRLIFA ASSLHNGVPS RFSGSGSGTK FTLTISSLQP EDLGTYYCL QYYSDPYTFG QGTKVEIKRT VAAPSVFIFP PSEDQVKSGT VSVVCLLNNF YPREASVKWK VDGALKTGNS Q ESVTEQDS ...String: DIQMTQSPSS LSASVGDRVT ITCRASQGID NYLNWYQQKP GKAPKRLIFA ASSLHNGVPS RFSGSGSGTK FTLTISSLQP EDLGTYYCL QYYSDPYTFG QGTKVEIKRT VAAPSVFIFP PSEDQVKSGT VSVVCLLNNF YPREASVKWK VDGALKTGNS Q ESVTEQDS KDNTYSLSST LTLSSTEYQS HKVYACEVTH QGLSSPVTKS FNRGEC |
-Macromolecule #8: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 8 / Number of copies: 27 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: PBS |
| Vitrification | Cryogen name: ETHANE |
| Details | SIV E660.CR54 SOS-2P with ITS92.02 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average exposure time: 2.0 sec. / Average electron dose: 51.16 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
|---|---|
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.32 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 32347 |
 Movie
Movie Controller
Controller












 Z
Z Y
Y X
X