[English] 日本語
 Yorodumi
Yorodumi- EMDB-27387: Cryo-EM structure of the zebrafish two pore domain K+ channel TRE... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the zebrafish two pore domain K+ channel TREK1 (K2P2.1) in DDM/POPA mixed micelles | |||||||||
 Map data Map data | drTREK1 in DDM/POPA mixed micelles full map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology | :  Function and homology information Function and homology information | |||||||||
| Biological species |   Danio rerio (zebrafish) Danio rerio (zebrafish) | |||||||||
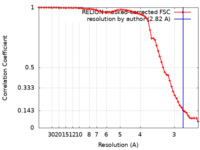
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.82 Å cryo EM / Resolution: 2.82 Å | |||||||||
 Authors Authors | Schmidpeter PAM / Nimigean CM / Riegelhaupt PM | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Membrane phospholipids control gating of the mechanosensitive potassium leak channel TREK1. Authors: Philipp A M Schmidpeter / John T Petroff / Leila Khajoueinejad / Aboubacar Wague / Cheryl Frankfater / Wayland W L Cheng / Crina M Nimigean / Paul M Riegelhaupt /  Abstract: Tandem pore domain (K2P) potassium channels modulate resting membrane potentials and shape cellular excitability. For the mechanosensitive subfamily of K2Ps, the composition of phospholipids within ...Tandem pore domain (K2P) potassium channels modulate resting membrane potentials and shape cellular excitability. For the mechanosensitive subfamily of K2Ps, the composition of phospholipids within the bilayer strongly influences channel activity. To examine the molecular details of K2P lipid modulation, we solved cryo-EM structures of the TREK1 K2P channel bound to either the anionic lipid phosphatidic acid (PA) or the zwitterionic lipid phosphatidylethanolamine (PE). At the extracellular face of TREK1, a PA lipid inserts its hydrocarbon tail into a pocket behind the selectivity filter, causing a structural rearrangement that recapitulates mutations and pharmacology known to activate TREK1. At the cytoplasmic face, PA and PE lipids compete to modulate the conformation of the TREK1 TM4 gating helix. Our findings demonstrate two distinct pathways by which anionic lipids enhance TREK1 activity and provide a framework for a model that integrates lipid gating with the effects of other mechanosensitive K2P modulators. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27387.map.gz emd_27387.map.gz | 19.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27387-v30.xml emd-27387-v30.xml emd-27387.xml emd-27387.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27387_fsc.xml emd_27387_fsc.xml | 6.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_27387.png emd_27387.png | 1.4 MB | ||
| Masks |  emd_27387_msk_1.map emd_27387_msk_1.map | 27 MB |  Mask map Mask map | |
| Others |  emd_27387_half_map_1.map.gz emd_27387_half_map_1.map.gz emd_27387_half_map_2.map.gz emd_27387_half_map_2.map.gz | 19.8 MB 19.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27387 http://ftp.pdbj.org/pub/emdb/structures/EMD-27387 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27387 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27387 | HTTPS FTP |
-Related structure data
| Related structure data |  8de8MC  8de7C  8de9C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27387.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27387.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | drTREK1 in DDM/POPA mixed micelles full map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.278 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27387_msk_1.map emd_27387_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
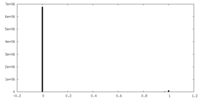
| Density Histograms |
-Half map: drTREK1 in DDM/POPA mixed micelles halfmap1
| File | emd_27387_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | drTREK1 in DDM/POPA mixed micelles halfmap1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
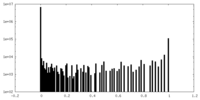
| Density Histograms |
-Half map: drTREK1 in DDM/POPA mixed micelles halfmap2
| File | emd_27387_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | drTREK1 in DDM/POPA mixed micelles halfmap2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
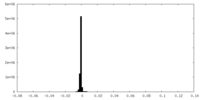
| Density Histograms |
- Sample components
Sample components
-Entire : Potassium channel subfamily K member 2
| Entire | Name: Potassium channel subfamily K member 2 |
|---|---|
| Components |
|
-Supramolecule #1: Potassium channel subfamily K member 2
| Supramolecule | Name: Potassium channel subfamily K member 2 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Danio rerio (zebrafish) Danio rerio (zebrafish) |
-Macromolecule #1: Potassium channel, subfamily K, member 2a
| Macromolecule | Name: Potassium channel, subfamily K, member 2a / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Danio rerio (zebrafish) Danio rerio (zebrafish) |
| Molecular weight | Theoretical: 35.559383 KDa |
| Recombinant expression | Organism:   Komagataella pastoris (fungus) Komagataella pastoris (fungus) |
| Sequence | String: MAAPDLLDPK SATHNTKPRL SFSSKPIVYN SGDDCESITT VMKWKTVLAI FLLVVLYLII GATVFKALEQ PEEGLQKYRI IQEKIDFLS MHTCVQTSEL EDLVKQVVLA IRAGVNPSGH PSQESSMWDL SSSFFFAGTV ITTIGFGNVS PHTEGGRIFC I IYALLGIP ...String: MAAPDLLDPK SATHNTKPRL SFSSKPIVYN SGDDCESITT VMKWKTVLAI FLLVVLYLII GATVFKALEQ PEEGLQKYRI IQEKIDFLS MHTCVQTSEL EDLVKQVVLA IRAGVNPSGH PSQESSMWDL SSSFFFAGTV ITTIGFGNVS PHTEGGRIFC I IYALLGIP LFGFLLAGVG DQLGTIFGKG IAKVEKMFVK WNVSQTKIRV TSTVLFILFG CLLFVALPAL IFQHIEGWSA LE SIYFVVI TLTTIGFGDF VAGGSEIEYL DYYKPIVWFW ILVGLAYFAA VLSMIGDWLR VISKKTKEEV GEFRAHAAEW TAN V |
-Macromolecule #2: (2R)-1-(hexadecanoyloxy)-3-(phosphonooxy)propan-2-yl (9Z)-octadec...
| Macromolecule | Name: (2R)-1-(hexadecanoyloxy)-3-(phosphonooxy)propan-2-yl (9Z)-octadec-9-enoate type: ligand / ID: 2 / Number of copies: 4 / Formula: D21 |
|---|---|
| Molecular weight | Theoretical: 674.929 Da |
| Chemical component information |  ChemComp-D21: |
-Macromolecule #3: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 3 / Number of copies: 4 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 150mM KCl, 20mM TRIS, 0.25mM DDM, 0.1mg/ml POPA |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 80 sec. / Pretreatment - Pressure: 0.03 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 294 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.3 µm / Nominal magnification: 105000 Bright-field microscopy / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.3 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Software | Name: Leginon |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 63.32 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Software | Name:  UCSF Chimera (ver. 1.14.0) UCSF Chimera (ver. 1.14.0)Details: Model was first docked into density using Chimera Fit in Map tool |
|---|---|
| Output model |  PDB-8de8: |
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X