[English] 日本語
 Yorodumi
Yorodumi- EMDB-27025: Cryo-EM structure of Human 15-PGDH in complex with small molecule... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Human 15-PGDH in complex with small molecule SW222746 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information 15-hydroxyprostaglandin dehydrogenase (NAD+) / 15-hydroxyprostaglandin dehydrogenase (NAD+) /  15-hydroxyicosatetraenoate dehydrogenase / regulation of prostaglandin catabolic process / ductus arteriosus closure / 15-hydroxyicosatetraenoate dehydrogenase / regulation of prostaglandin catabolic process / ductus arteriosus closure /  ovulation / thrombin-activated receptor signaling pathway / ovulation / thrombin-activated receptor signaling pathway /  prostaglandin E receptor activity / prostaglandin E receptor activity /  15-hydroxyprostaglandin dehydrogenase (NAD+) activity / Synthesis of Lipoxins (LX) / 15-hydroxyprostaglandin dehydrogenase (NAD+) activity / Synthesis of Lipoxins (LX) /  parturition ... parturition ... 15-hydroxyprostaglandin dehydrogenase (NAD+) / 15-hydroxyprostaglandin dehydrogenase (NAD+) /  15-hydroxyicosatetraenoate dehydrogenase / regulation of prostaglandin catabolic process / ductus arteriosus closure / 15-hydroxyicosatetraenoate dehydrogenase / regulation of prostaglandin catabolic process / ductus arteriosus closure /  ovulation / thrombin-activated receptor signaling pathway / ovulation / thrombin-activated receptor signaling pathway /  prostaglandin E receptor activity / prostaglandin E receptor activity /  15-hydroxyprostaglandin dehydrogenase (NAD+) activity / Synthesis of Lipoxins (LX) / 15-hydroxyprostaglandin dehydrogenase (NAD+) activity / Synthesis of Lipoxins (LX) /  parturition / lipoxygenase pathway / Biosynthesis of D-series resolvins / Biosynthesis of E-series 18(S)-resolvins / Synthesis of Prostaglandins (PG) and Thromboxanes (TX) / parturition / lipoxygenase pathway / Biosynthesis of D-series resolvins / Biosynthesis of E-series 18(S)-resolvins / Synthesis of Prostaglandins (PG) and Thromboxanes (TX) /  Oxidoreductases; Acting on the CH-OH group of donors; With NAD+ or NADP+ as acceptor / oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor / prostaglandin metabolic process / negative regulation of cell cycle / Oxidoreductases; Acting on the CH-OH group of donors; With NAD+ or NADP+ as acceptor / oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor / prostaglandin metabolic process / negative regulation of cell cycle /  NAD+ binding / positive regulation of vascular associated smooth muscle cell proliferation / transforming growth factor beta receptor signaling pathway / NAD+ binding / positive regulation of vascular associated smooth muscle cell proliferation / transforming growth factor beta receptor signaling pathway /  kidney development / female pregnancy / NAD binding / response to estradiol / basolateral plasma membrane / response to ethanol / response to lipopolysaccharide / positive regulation of apoptotic process / extracellular exosome / kidney development / female pregnancy / NAD binding / response to estradiol / basolateral plasma membrane / response to ethanol / response to lipopolysaccharide / positive regulation of apoptotic process / extracellular exosome /  nucleoplasm / identical protein binding / nucleoplasm / identical protein binding /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.9 Å cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Huang W / Taylor DJ | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Small molecule inhibitors of 15-PGDH exploit a physiologic induced-fit closing system. Authors: Wei Huang / Hongyun Li / Janna Kiselar / Stephen P Fink / Sagar Regmi / Alexander Day / Yiyuan Yuan / Mark Chance / Joseph M Ready / Sanford D Markowitz / Derek J Taylor /  Abstract: 15-prostaglandin dehydrogenase (15-PGDH) is a negative regulator of tissue stem cells that acts via enzymatic activity of oxidizing and degrading PGE2, and related eicosanoids, that support stem ...15-prostaglandin dehydrogenase (15-PGDH) is a negative regulator of tissue stem cells that acts via enzymatic activity of oxidizing and degrading PGE2, and related eicosanoids, that support stem cells during tissue repair. Indeed, inhibiting 15-PGDH markedly accelerates tissue repair in multiple organs. Here we have used cryo-electron microscopy to solve the solution structure of native 15-PGDH and of 15-PGDH individually complexed with two distinct chemical inhibitors. These structures identify key 15-PGDH residues that mediate binding to both classes of inhibitors. Moreover, we identify a dynamic 15-PGDH lid domain that closes around the inhibitors, and that is likely fundamental to the physiologic 15-PGDH enzymatic mechanism. We furthermore identify two key residues, F185 and Y217, that act as hinges to regulate lid closing, and which both inhibitors exploit to capture the lid in the closed conformation, thus explaining their sub-nanomolar binding affinities. These findings provide the basis for further development of 15-PGDH targeted drugs as therapeutics for regenerative medicine. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27025.map.gz emd_27025.map.gz | 32.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27025-v30.xml emd-27025-v30.xml emd-27025.xml emd-27025.xml | 15.1 KB 15.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27025.png emd_27025.png | 104.4 KB | ||
| Others |  emd_27025_half_map_1.map.gz emd_27025_half_map_1.map.gz emd_27025_half_map_2.map.gz emd_27025_half_map_2.map.gz | 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27025 http://ftp.pdbj.org/pub/emdb/structures/EMD-27025 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27025 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27025 | HTTPS FTP |
-Related structure data
| Related structure data |  8cwlMC  8cvnC  8fd8C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27025.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27025.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.02 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_27025_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_27025_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : HUMAN 15-PGDH IN COMPLEX WITH INHIBITOR
| Entire | Name: HUMAN 15-PGDH IN COMPLEX WITH INHIBITOR |
|---|---|
| Components |
|
-Supramolecule #1: HUMAN 15-PGDH IN COMPLEX WITH INHIBITOR
| Supramolecule | Name: HUMAN 15-PGDH IN COMPLEX WITH INHIBITOR / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: 15-hydroxyprostaglandin dehydrogenase [NAD(+)]
| Macromolecule | Name: 15-hydroxyprostaglandin dehydrogenase [NAD(+)] / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number:  15-hydroxyprostaglandin dehydrogenase (NAD+) 15-hydroxyprostaglandin dehydrogenase (NAD+) |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 27.747906 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MVNGKVALVT GAAQGIGRAF AEALLLKGAK VALVDWNLEA GVQCKAALDE QFEPQKTLFI QCDVADQQQL RDTFRKVVDH FGRLDILVN NAGVNNEKNW EKTLQINLVS VISGTYLGLD YMSKQNGGEG GIIINMSSLA GLMPVAQQPV YCASKHGIVG F TRSAALAA ...String: MVNGKVALVT GAAQGIGRAF AEALLLKGAK VALVDWNLEA GVQCKAALDE QFEPQKTLFI QCDVADQQQL RDTFRKVVDH FGRLDILVN NAGVNNEKNW EKTLQINLVS VISGTYLGLD YMSKQNGGEG GIIINMSSLA GLMPVAQQPV YCASKHGIVG F TRSAALAA NLMNSGVRLN AICPGFVNTA ILESIEKEEN MGQYIEYKDH IKDMIKYYGI LDPPLIANGL ITLIEDDALN GA IMKITTS KGIHFQDY |
-Macromolecule #2: 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE
| Macromolecule | Name: 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE / type: ligand / ID: 2 / Number of copies: 2 / Formula: NAI |
|---|---|
| Molecular weight | Theoretical: 665.441 Da |
| Chemical component information |  ChemComp-NAI: |
-Macromolecule #3: 2-methyl-6-[7-(piperidine-1-carbonyl)quinoxalin-2-yl]isoquinolin-...
| Macromolecule | Name: 2-methyl-6-[7-(piperidine-1-carbonyl)quinoxalin-2-yl]isoquinolin-1(2H)-one type: ligand / ID: 3 / Number of copies: 2 / Formula: RLD |
|---|---|
| Molecular weight | Theoretical: 398.457 Da |
| Chemical component information | 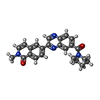 ChemComp-RLD: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.25 µm Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.25 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 1.08 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing #1
Image processing #1
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
|---|---|
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 361366 |
| Image processing ID | 1 |
- Image processing #2
Image processing #2
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
|---|---|
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 361366 |
| Image processing ID | 2 |
 Movie
Movie Controller
Controller










 Z
Z Y
Y X
X

















