+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24457 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mycobacterial CIII2CIV2 supercomplex, Telacebec (Q203) bound | ||||||||||||
 Map data Map data | Mycobacterium CIII2CIV2 supercomplex, telacebec bound. | ||||||||||||
 Sample Sample |
| ||||||||||||
| Function / homology |  Function and homology information Function and homology informationaerobic electron transport chain /  cytochrome-c oxidase / quinol-cytochrome-c reductase / cytochrome-c oxidase / quinol-cytochrome-c reductase /  ubiquinol-cytochrome-c reductase activity / ubiquinol-cytochrome-c reductase activity /  cytochrome-c oxidase activity / cytochrome-c oxidase activity /  superoxide dismutase / superoxide dismutase /  superoxide dismutase activity / oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen / superoxide dismutase activity / oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen /  respirasome / respiratory electron transport chain ...aerobic electron transport chain / respirasome / respiratory electron transport chain ...aerobic electron transport chain /  cytochrome-c oxidase / quinol-cytochrome-c reductase / cytochrome-c oxidase / quinol-cytochrome-c reductase /  ubiquinol-cytochrome-c reductase activity / ubiquinol-cytochrome-c reductase activity /  cytochrome-c oxidase activity / cytochrome-c oxidase activity /  superoxide dismutase / superoxide dismutase /  superoxide dismutase activity / oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen / superoxide dismutase activity / oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen /  respirasome / respiratory electron transport chain / respirasome / respiratory electron transport chain /  monooxygenase activity / 2 iron, 2 sulfur cluster binding / iron ion binding / copper ion binding / monooxygenase activity / 2 iron, 2 sulfur cluster binding / iron ion binding / copper ion binding /  heme binding / heme binding /  membrane / membrane /  metal ion binding / metal ion binding /  plasma membrane plasma membraneSimilarity search - Function | ||||||||||||
| Biological species |  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) | ||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.0 Å cryo EM / Resolution: 3.0 Å | ||||||||||||
 Authors Authors | Di Trani JM / Yanofsky DJ / Rubinstein JL | ||||||||||||
| Funding support |  Canada, Canada,  Sweden, 3 items Sweden, 3 items
| ||||||||||||
 Citation Citation |  Journal: Elife / Year: 2021 Journal: Elife / Year: 2021Title: Structure of mycobacterial CIIICIV respiratory supercomplex bound to the tuberculosis drug candidate telacebec (Q203). Authors: David J Yanofsky / Justin M Di Trani / Sylwia Król / Rana Abdelaziz / Stephanie A Bueler / Peter Imming / Peter Brzezinski / John L Rubinstein /    Abstract: The imidazopyridine telacebec, also known as Q203, is one of only a few new classes of compounds in more than 50 years with demonstrated antituberculosis activity in humans. Telacebec inhibits the ...The imidazopyridine telacebec, also known as Q203, is one of only a few new classes of compounds in more than 50 years with demonstrated antituberculosis activity in humans. Telacebec inhibits the mycobacterial respiratory supercomplex composed of complexes III and IV (CIIICIV). In mycobacterial electron transport chains, CIIICIV replaces canonical CIII and CIV, transferring electrons from the intermediate carrier menaquinol to the final acceptor, molecular oxygen, while simultaneously transferring protons across the inner membrane to power ATP synthesis. We show that telacebec inhibits the menaquinol:oxygen oxidoreductase activity of purified CIIICIV at concentrations similar to those needed to inhibit electron transfer in mycobacterial membranes and growth in culture. We then used electron cryomicroscopy (cryoEM) to determine structures of CIIICIV both in the presence and absence of telacebec. The structures suggest that telacebec prevents menaquinol oxidation by blocking two different menaquinol binding modes to prevent CIIICIV activity. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24457.map.gz emd_24457.map.gz | 102.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24457-v30.xml emd-24457-v30.xml emd-24457.xml emd-24457.xml | 27 KB 27 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24457.png emd_24457.png | 77.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24457 http://ftp.pdbj.org/pub/emdb/structures/EMD-24457 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24457 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24457 | HTTPS FTP |
-Related structure data
| Related structure data |  7rh7MC  7rh5C  7rh6C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24457.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24457.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mycobacterium CIII2CIV2 supercomplex, telacebec bound. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Respiratory Complex CIII2CIV2SOD2 from Mycobacterium smegmatis
+Supramolecule #1: Respiratory Complex CIII2CIV2SOD2 from Mycobacterium smegmatis
+Macromolecule #1: LpqE protein
+Macromolecule #2: Cytochrome aa3 subunit 2
+Macromolecule #3: Cytochrome c oxidase subunit 1
+Macromolecule #4: Cytochrome aa3 subunit 3
+Macromolecule #5: Cytochrome c oxidase polypeptide 4
+Macromolecule #6: Cytochrome c oxidase subunit CtaJ
+Macromolecule #7: Uncharacterized protein MSMEG_4692/MSMEI_4575
+Macromolecule #8: Conserved transmembrane protein
+Macromolecule #9: Superoxide dismutase [Cu-Zn]
+Macromolecule #10: Cytochrome bc1 complex cytochrome c subunit
+Macromolecule #11: Cytochrome bc1 complex cytochrome b subunit
+Macromolecule #12: Cytochrome bc1 complex Rieske iron-sulfur subunit
+Macromolecule #13: (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate
+Macromolecule #14: PALMITIC ACID
+Macromolecule #15: COPPER (II) ION
+Macromolecule #16: HEME-A
+Macromolecule #17: CARDIOLIPIN
+Macromolecule #18: (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)...
+Macromolecule #19: MENAQUINONE-9
+Macromolecule #20: HEME C
+Macromolecule #21: (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3...
+Macromolecule #22: PROTOPORPHYRIN IX CONTAINING FE
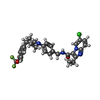
+Macromolecule #23: 6-chloranyl-2-ethyl-N-[[4-[4-[4-(trifluoromethyloxy)phenyl]piperi...
+Macromolecule #24: FE2/S2 (INORGANIC) CLUSTER
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 43.5 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
|---|---|
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 70818 |
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-7rh7: |
 Movie
Movie Controller
Controller