[English] 日本語
 Yorodumi
Yorodumi- EMDB-24071: Cryo-EM structure of broadly neutralizing V2-apex-targeting antib... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of broadly neutralizing V2-apex-targeting antibody J038 in complex with HIV-1 Env | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information: / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / virus-mediated perturbation of host defense response / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell /  viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane ...: / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / virus-mediated perturbation of host defense response / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane ...: / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / virus-mediated perturbation of host defense response / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell /  viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /    Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /   Macaca mulatta (Rhesus monkey) Macaca mulatta (Rhesus monkey) | |||||||||
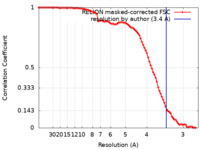
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.4 Å cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Zhou T / Gao F | |||||||||
 Citation Citation |  Journal: Adv Sci (Weinh) / Year: 2022 Journal: Adv Sci (Weinh) / Year: 2022Title: Development of Neutralization Breadth against Diverse HIV-1 by Increasing Ab-Ag Interface on V2. Authors: Nan Gao / Yanxin Gai / Lina Meng / Chu Wang / Wei Wang / Xiaojun Li / Tiejun Gu / Mark K Louder / Nicole A Doria-Rose / Kevin Wiehe / Alexandra F Nazzari / Adam S Olia / Jason Gorman / Reda ...Authors: Nan Gao / Yanxin Gai / Lina Meng / Chu Wang / Wei Wang / Xiaojun Li / Tiejun Gu / Mark K Louder / Nicole A Doria-Rose / Kevin Wiehe / Alexandra F Nazzari / Adam S Olia / Jason Gorman / Reda Rawi / Wenmin Wu / Clayton Smith / Htet Khant / Natalia de Val / Bin Yu / Junhong Luo / Haitao Niu / Yaroslav Tsybovsky / Huaxin Liao / Thomas B Kepler / Peter D Kwong / John R Mascola / Chuan Qin / Tongqing Zhou / Xianghui Yu / Feng Gao /   Abstract: Understanding maturation pathways of broadly neutralizing antibodies (bnAbs) against HIV-1 can be highly informative for HIV-1 vaccine development. A lineage of J038 bnAbs is now obtained from a long- ...Understanding maturation pathways of broadly neutralizing antibodies (bnAbs) against HIV-1 can be highly informative for HIV-1 vaccine development. A lineage of J038 bnAbs is now obtained from a long-term SHIV-infected macaque. J038 neutralizes 54% of global circulating HIV-1 strains. Its binding induces a unique "up" conformation for one of the V2 loops in the trimeric envelope glycoprotein and is heavily dependent on glycan, which provides nearly half of the binding surface. Their unmutated common ancestor neutralizes the autologous virus. Continuous maturation enhances neutralization potency and breadth of J038 lineage antibodies via expanding antibody-Env contact areas surrounding the core region contacted by germline-encoded residues. Developmental details and recognition features of J038 lineage antibodies revealed here provide a new pathway for elicitation and maturation of V2-targeting bnAbs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24071.map.gz emd_24071.map.gz | 5.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24071-v30.xml emd-24071-v30.xml emd-24071.xml emd-24071.xml | 23.2 KB 23.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24071_fsc.xml emd_24071_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_24071.png emd_24071.png | 155.1 KB | ||
| Others |  emd_24071_half_map_1.map.gz emd_24071_half_map_1.map.gz emd_24071_half_map_2.map.gz emd_24071_half_map_2.map.gz | 49.6 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24071 http://ftp.pdbj.org/pub/emdb/structures/EMD-24071 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24071 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24071 | HTTPS FTP |
-Related structure data
| Related structure data |  7mxdMC  7n28C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24071.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24071.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_24071_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
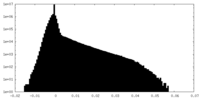
| Projections & Slices |
| ||||||||||||
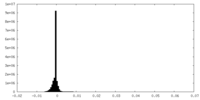
| Density Histograms |
-Half map: #2
| File | emd_24071_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
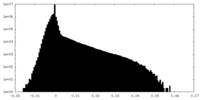
| Projections & Slices |
| ||||||||||||
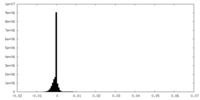
| Density Histograms |
- Sample components
Sample components
-Entire : A complex of HIV-1 Env with antibodies 3BNC117 and J038
| Entire | Name: A complex of HIV-1 Env with antibodies 3BNC117 and J038 |
|---|---|
| Components |
|
-Supramolecule #1: A complex of HIV-1 Env with antibodies 3BNC117 and J038
| Supramolecule | Name: A complex of HIV-1 Env with antibodies 3BNC117 and J038 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Envelope glycoprotein gp120
| Macromolecule | Name: Envelope glycoprotein gp120 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 54.23475 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AENLWVTVYY GVPVWKDADT TLFCASDAKA HETEAHNIWA THACVPTDPN PQEIYMENVT ENFNMWKNNM VEQMQEDIIS LWDQSLKPC VKLTPLCVTL SCTNVTLTNV NYTNNFPNIG NITDEVRNCS FNVTTEIRDK KQKVYALFYK LDIVQMENKN S YRLINCNT ...String: AENLWVTVYY GVPVWKDADT TLFCASDAKA HETEAHNIWA THACVPTDPN PQEIYMENVT ENFNMWKNNM VEQMQEDIIS LWDQSLKPC VKLTPLCVTL SCTNVTLTNV NYTNNFPNIG NITDEVRNCS FNVTTEIRDK KQKVYALFYK LDIVQMENKN S YRLINCNT SVCKQACPKI SFDPIPIHYC TPAGYAILKC NEKNFNGTGP CKNVSSVQCT HGIKPVVSTQ LLLNGSLAEG EI IIRSENL TNNAKTIIVH LNKSVEINCT RPSNNTRTSV TIGPGQVFYR TGDIIGDIRK AYCEINGTKW NETLKQVVGK LKE HFPNKT ISFQPPSGGD LEITMHHFNC RGEFFYCNTT QLFNSTWINS TTIKEYNDTI IYLPCKIKQI INMWQGVGQC MYAP PIRGK INCVSNITGI LLTRDGGDAN ATNDTETFRP GGGNIKDNWR SELYKYKVVQ IEPLGIAPTK CKRRVVERRR RRR |
-Macromolecule #2: 3BNC117 antibody heavy chain
| Macromolecule | Name: 3BNC117 antibody heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.656484 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLLQSGAA VTKPGASVRV SCEASGYNIR DYFIHWWRQA PGQGLQWVGW INPKTGQPNN PRQFQGRVSL TRHASWDFDT YSFYMDLKA LRSDDTAVYF CARQRSDYWD FDVWGSGTQV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA ...String: QVQLLQSGAA VTKPGASVRV SCEASGYNIR DYFIHWWRQA PGQGLQWVGW INPKTGQPNN PRQFQGRVSL TRHASWDFDT YSFYMDLKA LRSDDTAVYF CARQRSDYWD FDVWGSGTQV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA LTSGVHTFPA VLQSSGLYSL SSVVTVPSSS LGTQTYICNV NHKPSNTKVD KKVEPKSC |
-Macromolecule #3: 3BNC117 antibody light chain
| Macromolecule | Name: 3BNC117 antibody light chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.022658 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS LSASVGDTVT ITCQANGYLN WYQQRRGKAP KLLIYDGSKL ERGVPSRFSG RRWGQEYNLT INNLQPEDIA TYFCQVYEF VVPGTRLDLK RTVAAPSVFI FPPSDEQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG NSQESVTEQD S KDSTYSLS ...String: DIQMTQSPSS LSASVGDTVT ITCQANGYLN WYQQRRGKAP KLLIYDGSKL ERGVPSRFSG RRWGQEYNLT INNLQPEDIA TYFCQVYEF VVPGTRLDLK RTVAAPSVFI FPPSDEQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG NSQESVTEQD S KDSTYSLS STLTLSKADY EKHKVYACEV THQGLSSPVT KSFNRGEC |
-Macromolecule #4: J038 antibody heavy chain
| Macromolecule | Name: J038 antibody heavy chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Macaca mulatta (Rhesus monkey) Macaca mulatta (Rhesus monkey) |
| Molecular weight | Theoretical: 24.413338 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QLHLQESGPG LVRPSETLSL TCDVSGGAFN DAYCSWIRRF PGGGLEWIGR ISGRDGYVES NPALTGRVTM SIDATWKKIV LRLTSMTAS DTATYFCAGE TPEDDFGYYQ PYFKTWGQGL GVTVSSASTK GPSVFPLAPC SRSTSESTAA LGCLVKDYFP E PVTVSWNS ...String: QLHLQESGPG LVRPSETLSL TCDVSGGAFN DAYCSWIRRF PGGGLEWIGR ISGRDGYVES NPALTGRVTM SIDATWKKIV LRLTSMTAS DTATYFCAGE TPEDDFGYYQ PYFKTWGQGL GVTVSSASTK GPSVFPLAPC SRSTSESTAA LGCLVKDYFP E PVTVSWNS GSLTSGVHTF PAVLQSSGLY SLSSVVTVPS SSLGTQTYVC NVNHKPSNTK VDKRVEIKTC |
-Macromolecule #5: J038 antibody light chain
| Macromolecule | Name: J038 antibody light chain / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Macaca mulatta (Rhesus monkey) Macaca mulatta (Rhesus monkey) |
| Molecular weight | Theoretical: 23.187713 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQLIQSPSS VSASLGDRVT ITCRSTQGIG SDLAWYQATP GTAPKLLIYN SFALHKGVPS RFSGSGSGTE FSLTITGLQP EDFATYYCQ HYRRLPLTFG GGTNIEVKRA VAAPSVFIFP PSEDQVKSGT VSVVCLLNNF YPREASVKWK VDGVLKTGNS Q ESVTEQDS ...String: DIQLIQSPSS VSASLGDRVT ITCRSTQGIG SDLAWYQATP GTAPKLLIYN SFALHKGVPS RFSGSGSGTE FSLTITGLQP EDFATYYCQ HYRRLPLTFG GGTNIEVKRA VAAPSVFIFP PSEDQVKSGT VSVVCLLNNF YPREASVKWK VDGVLKTGNS Q ESVTEQDS KDNTYSLSST LTLSNTDYQS HNVYACEVTH QGLSSPVTKS FNRGEC |
-Macromolecule #6: Envelope glycoprotein gp41
| Macromolecule | Name: Envelope glycoprotein gp41 / type: protein_or_peptide / ID: 6 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.2185 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAMIFG FLGAAGSTMG AASNTLTVQA RQLLSGIVQQ QSNLPRAPEA QQHLLQLTVW GIKQLQARVL AVERYLEVQK FLGLWGCSG KIICCTAVPW NSTWSNKSFE QIWNNMTWIE WEREISNYTS QIYDILTESQ FQQDINEVDL LELD |
-Macromolecule #12: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 12 / Number of copies: 38 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #13: PHOSPHATE ION
| Macromolecule | Name: PHOSPHATE ION / type: ligand / ID: 13 / Number of copies: 3 / Formula: PO4 |
|---|---|
| Molecular weight | Theoretical: 94.971 Da |
| Chemical component information |  ChemComp-PO4: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 18000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 18000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-7mxd: |
 Movie
Movie Controller
Controller







 Z
Z Y
Y X
X