+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22896 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The Neck region of Phage XM1 (6-fold symmetry) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Vibrio phage XM1 (virus) Vibrio phage XM1 (virus) | |||||||||
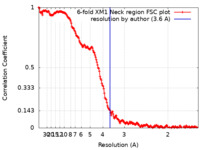
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.6 Å cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Wang Z / Klose T / Jiang W / Kuhn RJ | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Biorxiv / Year: 2021 Journal: Biorxiv / Year: 2021Title: Structure of Vibrio phage XM1, a simple contractile DNA injection machine Authors: Wang Z / Fokine A / Guo X / Jiang W / Rossmann MG / Kuhn RJ / Luo ZH / Klose T | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22896.map.gz emd_22896.map.gz | 51.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22896-v30.xml emd-22896-v30.xml emd-22896.xml emd-22896.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22896_fsc.xml emd_22896_fsc.xml | 18.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_22896.png emd_22896.png | 142.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22896 http://ftp.pdbj.org/pub/emdb/structures/EMD-22896 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22896 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22896 | HTTPS FTP |
-Related structure data
| Related structure data |  7kjkMC  7kh1C  7klnC  7kmxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22896.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22896.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.81 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Vibrio phage XM1
| Entire | Name:  Vibrio phage XM1 (virus) Vibrio phage XM1 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Vibrio phage XM1
| Supramolecule | Name: Vibrio phage XM1 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2748688 / Sci species name: Vibrio phage XM1 / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Vibrio rotiferianus (bacteria) Vibrio rotiferianus (bacteria) |
| Virus shell | Shell ID: 1 / Diameter: 640.0 Å / T number (triangulation number): 7 |
-Macromolecule #1: Tail terminator protein
| Macromolecule | Name: Tail terminator protein / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio phage XM1 (virus) Vibrio phage XM1 (virus) |
| Molecular weight | Theoretical: 18.236037 KDa |
| Sequence | String: MSQSIINVAR YIRDLLDYDE NLIQFDRKNT QQSDTVTGYI VVNGSGVQNV LSHGSSYDGD AEIMEYSKSE SRLITLEFYG SDAYENAEL FSLLNQSQKA KEVSRGLGLT IYNVSQATDV KQLLGYQYGN RVHVDFNIQY CPSVYVETLR VDASEFEILV D D |
-Macromolecule #2: Collar spike protein
| Macromolecule | Name: Collar spike protein / type: protein_or_peptide / ID: 2 / Number of copies: 18 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio phage XM1 (virus) Vibrio phage XM1 (virus) |
| Molecular weight | Theoretical: 90.692562 KDa |
| Sequence | String: MSIQVTGIIE GPLGTPSPGI TIRVVSKISY RNTYRLSTED YVTTTGGAYD FQLNEGFHKI LIRYRGASSF TKLGDVSVSD QTPSPITLI NLLESSSPKA LVVKLQELAD EASESAQSAS DDADQVTLDK QQVTLLAQQV SDDADQVALD RSATAQSEVN A AQSATTAQ ...String: MSIQVTGIIE GPLGTPSPGI TIRVVSKISY RNTYRLSTED YVTTTGGAYD FQLNEGFHKI LIRYRGASSF TKLGDVSVSD QTPSPITLI NLLESSSPKA LVVKLQELAD EASESAQSAS DDADQVTLDK QQVTLLAQQV SDDADQVALD RSATAQSEVN A AQSATTAQ QSADDAAAIV PNLQSQIDDK IDKTQSIIPD VDALFSLGRH EINPTNNNIP GLSGTRSLLH IQNNLGGFQL AA RTVGSEI HVRSADTNSG NIGPWERLYT TGYRPSSYDL VVKDKAEALS VSHQEGLIIG IESCNGARFV SVLPSDSGGL YLG SLPNGN KLRHIPLDGF SLKTIRTSDF EINSGSDVSQ FLIDASTAGY GQVYIDNDCE VTQLCDITGV KYLESLNGAE MLID DLNGR VKCDTDGFEV KGIVFKKSGD YSNTTFEAYL LETNGEKSLA SRCQFVGQWA GYKLQGNDSS ADLCFFSDGT FMLRV GEGV SSCNINRCFF TRGTRRFTEG EQQGDGIKLT AQSSTQGYNE NIIISDCIFY DTYRDCIDGY VGANRLIVKG SIMLNW GAH ALDFKSRVDD NSESPDGGPL TRNVLVDGCM IWKDVVRPDF DLTLAAVVMS VIPGALPVPT SEADVRQNGL RGMELKN TL IAYKGNGSLI SAASTWRTVI DGCTLTDLGQ IGQIIYAPNS WHLKLINSTV NIDPSNINNV KFSSAMNNAE VNDNTFNR L LISTDGMIRT SIKRNKFNGE GVLSRGLNIA SEEVSAVSNE FENFTSFGAQ IFNSASKCRF DFNEIRNCPA PLGIENPSV NTYHSVRGNI SMGSGAFTGL NRITDNGGVA NGNDHVPYS |
-Macromolecule #3: Head completion protein
| Macromolecule | Name: Head completion protein / type: protein_or_peptide / ID: 3 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio phage XM1 (virus) Vibrio phage XM1 (virus) |
| Molecular weight | Theoretical: 13.045932 KDa |
| Sequence | String: MLPNMRSALK MFEQSVLLKS VETIRVDFVD DIIITATPIR AVVQVADKKK LNLDSLDWSK QYIWVHSGSK MEIGQFIEWH GKDFKLVAA GDDYSDYGYN AWYGEETLKP VLVSS |
-Macromolecule #4: Tail tube protein
| Macromolecule | Name: Tail tube protein / type: protein_or_peptide / ID: 4 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio phage XM1 (virus) Vibrio phage XM1 (virus) |
| Molecular weight | Theoretical: 15.665332 KDa |
| Sequence | String: MSVIVYRNNQ STLTLNGYTF QHLYQGAALV LTPVNAKTAR TNSINGGVSI SGRVDGGVHT LAIMVQKHSP DDKFLNDAKN SQEPVVFDG SMKRAYTESG TLKKATTTLE TGSITTQPTK TDNNQDPDDS RTYVIEFRNS VETF |
-Macromolecule #5: Tail sheath protein
| Macromolecule | Name: Tail sheath protein / type: protein_or_peptide / ID: 5 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio phage XM1 (virus) Vibrio phage XM1 (virus) |
| Molecular weight | Theoretical: 53.006762 KDa |
| Sequence | String: MASISEVIRV SLQQEGRAIA PDNMNAVGII TGNQGVLSTA DRYRIYRTAA AVASDFGASS QESAFANTFF DTTPNPISAG GVLVIGYWR SASETVAATS ATLVSEQTSE SVLIPLLNAI NDGSFTITVD GGTEQEVTAL DFTGVSELSE VATILNSAIT G ATVSEDNG ...String: MASISEVIRV SLQQEGRAIA PDNMNAVGII TGNQGVLSTA DRYRIYRTAA AVASDFGASS QESAFANTFF DTTPNPISAG GVLVIGYWR SASETVAATS ATLVSEQTSE SVLIPLLNAI NDGSFTITVD GGTEQEVTAL DFTGVSELSE VATILNSAIT G ATVSEDNG YFKVTSSTTG ATSLLSYLGV ATSGTDISAV LGMNSESGAV LTQGTDQVVL PAETKLEGIT AIKSEVNIKG AM FIDQILD ADIPGIASFA GANNMLVYEV FDTGYLSKNV SNPVWAVKLA GQSNFRCLLS KSGNRKFAAT YMARMHTVLF SGQ NTAITM QLKELSVTAE EYTDTEIANA KTVGLDLLTT IKNEQALLTS GANDFCDNVY NLEAFRDEIQ TNNYNLLKTT STKI PQTDP GMDTIEDDTE KTCEKYVRNG VFAPGTWTRS DFFGDRQQFV DAIAQKGYYV LIGDLADQTT AERQSRVSPV IQIAV KNAG AVHEEDIIIS VNL |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 50 mM Tris, pH 7.5, 100 mM NaCl, 8 mM MgSO4 |
|---|---|
| Grid | Model: PELCO Ultrathin Carbon with Lacey Carbon / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 298 K / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: DIRECT ELECTRON DE-16 (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 30.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller