+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bacillus subtilis MutS2-collided disome complex (stalled 70S) | |||||||||
 Map data Map data | Focus refined map filtered at FSC resolution | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Muts2 /  collision / disome / splitting / collision / disome / splitting /  quality control / quality control /  RIBOSOME RIBOSOME | |||||||||
| Function / homology |  Function and homology information Function and homology informationmismatched DNA binding / negative regulation of DNA recombination / ATP-dependent DNA damage sensor activity /  mismatch repair / large ribosomal subunit / small ribosomal subunit / mismatch repair / large ribosomal subunit / small ribosomal subunit /  transferase activity / transferase activity /  endonuclease activity / endonuclease activity /  tRNA binding / tRNA binding /  Hydrolases; Acting on ester bonds ...mismatched DNA binding / negative regulation of DNA recombination / ATP-dependent DNA damage sensor activity / Hydrolases; Acting on ester bonds ...mismatched DNA binding / negative regulation of DNA recombination / ATP-dependent DNA damage sensor activity /  mismatch repair / large ribosomal subunit / small ribosomal subunit / mismatch repair / large ribosomal subunit / small ribosomal subunit /  transferase activity / transferase activity /  endonuclease activity / endonuclease activity /  tRNA binding / tRNA binding /  Hydrolases; Acting on ester bonds / Hydrolases; Acting on ester bonds /  rRNA binding / rRNA binding /  ribosome / structural constituent of ribosome / ribosome / structural constituent of ribosome /  translation / translation /  ribonucleoprotein complex / ribonucleoprotein complex /  mRNA binding / mRNA binding /  ATP hydrolysis activity / zinc ion binding / ATP hydrolysis activity / zinc ion binding /  ATP binding / ATP binding /  metal ion binding / metal ion binding /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Bacillus subtilis (bacteria) Bacillus subtilis (bacteria) | |||||||||
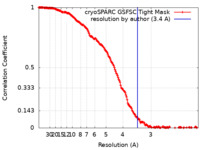
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.4 Å cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Park E / Mackens-Kiani T / Berhane R / Esser H / Erdenebat C / Burroughs AM / Berninghausen O / Aravind L / Beckmann R / Green R / Buskirk AR | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2024 Journal: EMBO J / Year: 2024Title: B. subtilis MutS2 splits stalled ribosomes into subunits without mRNA cleavage. Authors: Esther N Park / Timur Mackens-Kiani / Rebekah Berhane / Hanna Esser / Chimeg Erdenebat / A Maxwell Burroughs / Otto Berninghausen / L Aravind / Roland Beckmann / Rachel Green / Allen R Buskirk /   Abstract: Stalled ribosomes are rescued by pathways that recycle the ribosome and target the nascent polypeptide for degradation. In E. coli, these pathways are triggered by ribosome collisions through the ...Stalled ribosomes are rescued by pathways that recycle the ribosome and target the nascent polypeptide for degradation. In E. coli, these pathways are triggered by ribosome collisions through the recruitment of SmrB, a nuclease that cleaves the mRNA. In B. subtilis, the related protein MutS2 was recently implicated in ribosome rescue. Here we show that MutS2 is recruited to collisions by its SMR and KOW domains, and we reveal the interaction of these domains with collided ribosomes by cryo-EM. Using a combination of in vivo and in vitro approaches, we show that MutS2 uses its ABC ATPase activity to split ribosomes, targeting the nascent peptide for degradation through the ribosome quality control pathway. However, unlike SmrB, which cleaves mRNA in E. coli, we see no evidence that MutS2 mediates mRNA cleavage or promotes ribosome rescue by tmRNA. These findings clarify the biochemical and cellular roles of MutS2 in ribosome rescue in B. subtilis and raise questions about how these pathways function differently in diverse bacteria. #1:  Journal: Biorxiv / Year: 2023 Journal: Biorxiv / Year: 2023Title: An evolutionarily conserved strategy for ribosome binding and inhibition by beta-coronavirus non-structural protein 1 Authors: Maurina SF / O'Sullivan JP / Sharma G / Rodriguez DCP / MacFadden A / Cendali F / Henen MA / Kieft JS / Glasgow A / Steckelberg A | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18558.map.gz emd_18558.map.gz | 239.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18558-v30.xml emd-18558-v30.xml emd-18558.xml emd-18558.xml | 69.3 KB 69.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18558_fsc.xml emd_18558_fsc.xml | 16.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_18558.png emd_18558.png | 141.5 KB | ||
| Filedesc metadata |  emd-18558.cif.gz emd-18558.cif.gz | 14 KB | ||
| Others |  emd_18558_half_map_1.map.gz emd_18558_half_map_1.map.gz emd_18558_half_map_2.map.gz emd_18558_half_map_2.map.gz | 443.1 MB 443.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18558 http://ftp.pdbj.org/pub/emdb/structures/EMD-18558 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18558 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18558 | HTTPS FTP |
-Related structure data
| Related structure data |  8qppMC  8r55C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18558.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18558.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focus refined map filtered at FSC resolution | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_18558_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_18558_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : MutS2-disome complex from B. subtilis, focus refined, stalled 70S
+Supramolecule #1: MutS2-disome complex from B. subtilis, focus refined, stalled 70S
+Macromolecule #1: 50S ribosomal protein L32
+Macromolecule #2: Large ribosomal subunit protein bL33
+Macromolecule #3: Large ribosomal subunit protein bL34
+Macromolecule #4: Large ribosomal subunit protein bL35
+Macromolecule #5: 50S ribosomal protein L36
+Macromolecule #6: Large ribosomal subunit protein bL31
+Macromolecule #7: 30S ribosomal protein S2
+Macromolecule #8: 30S ribosomal protein S3
+Macromolecule #9: 30S ribosomal protein S4
+Macromolecule #10: 30S ribosomal protein S5
+Macromolecule #11: 30S ribosomal protein S6
+Macromolecule #12: 30S ribosomal protein S7
+Macromolecule #13: 30S ribosomal protein S8
+Macromolecule #14: 30S ribosomal protein S9
+Macromolecule #15: 30S ribosomal protein S10
+Macromolecule #16: 30S ribosomal protein S11
+Macromolecule #17: 30S ribosomal protein S12
+Macromolecule #18: 30S ribosomal protein S13
+Macromolecule #19: 30S ribosomal protein S14 type Z
+Macromolecule #20: 30S ribosomal protein S15
+Macromolecule #21: 30S ribosomal protein S16
+Macromolecule #22: 30S ribosomal protein S17
+Macromolecule #23: 30S ribosomal protein S18
+Macromolecule #24: 30S ribosomal protein S19
+Macromolecule #25: 30S ribosomal protein S20
+Macromolecule #28: Endonuclease MutS2
+Macromolecule #30: Large ribosomal subunit protein uL2
+Macromolecule #31: Large ribosomal subunit protein uL3
+Macromolecule #32: Large ribosomal subunit protein uL4
+Macromolecule #33: Large ribosomal subunit protein uL5
+Macromolecule #34: Large ribosomal subunit protein uL6
+Macromolecule #35: Large ribosomal subunit protein uL13
+Macromolecule #36: 50S ribosomal protein L14
+Macromolecule #37: 50S ribosomal protein L15
+Macromolecule #38: Large ribosomal subunit protein uL16
+Macromolecule #39: Large ribosomal subunit protein bL17
+Macromolecule #40: 50S ribosomal protein L18
+Macromolecule #41: 50S ribosomal protein L19
+Macromolecule #42: Large ribosomal subunit protein bL20
+Macromolecule #43: Large ribosomal subunit protein bL21
+Macromolecule #44: Large ribosomal subunit protein uL22
+Macromolecule #45: Large ribosomal subunit protein uL23
+Macromolecule #46: Large ribosomal subunit protein uL24
+Macromolecule #47: Large ribosomal subunit protein bL27
+Macromolecule #48: Large ribosomal subunit protein bL28
+Macromolecule #49: Large ribosomal subunit protein uL29
+Macromolecule #50: Large ribosomal subunit protein uL30
+Macromolecule #51: 50S ribosomal protein L9
+Macromolecule #26: tRNA (77-MER)
+Macromolecule #27: mRNA (33-MER)
+Macromolecule #29: 5S rRNA (112-MER)
+Macromolecule #52: 23S ribosomal RNA
+Macromolecule #53: 16S rRNA (1533-MER)
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.4 µm Bright-field microscopy / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.4 µm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 43.6 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z
Z Y
Y X
X