+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Hydrogen-dependent CO2 reductase. | ||||||||||||||||||
 Map data Map data | 3.4 Angstrom Cryo-EM structure of HDCR complex. The data was processed using cryosparc software. | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology information formate dehydrogenase / formate metabolic process / formate dehydrogenase / formate metabolic process /  ferredoxin hydrogenase / formate dehydrogenase (NAD+) activity / ferredoxin hydrogenase / formate dehydrogenase (NAD+) activity /  ferredoxin hydrogenase activity / ferredoxin hydrogenase activity /  Oxidoreductases / molybdopterin cofactor binding / Oxidoreductases / molybdopterin cofactor binding /  iron-sulfur cluster binding / 4 iron, 4 sulfur cluster binding / iron-sulfur cluster binding / 4 iron, 4 sulfur cluster binding /  oxidoreductase activity ... oxidoreductase activity ... formate dehydrogenase / formate metabolic process / formate dehydrogenase / formate metabolic process /  ferredoxin hydrogenase / formate dehydrogenase (NAD+) activity / ferredoxin hydrogenase / formate dehydrogenase (NAD+) activity /  ferredoxin hydrogenase activity / ferredoxin hydrogenase activity /  Oxidoreductases / molybdopterin cofactor binding / Oxidoreductases / molybdopterin cofactor binding /  iron-sulfur cluster binding / 4 iron, 4 sulfur cluster binding / iron-sulfur cluster binding / 4 iron, 4 sulfur cluster binding /  oxidoreductase activity / iron ion binding / oxidoreductase activity / iron ion binding /  metal ion binding metal ion bindingSimilarity search - Function | ||||||||||||||||||
| Biological species |    Thermoanaerobacter kivui LKT-1 (bacteria) / Thermoanaerobacter kivui LKT-1 (bacteria) /    Thermoanaerobacter kivui (bacteria) Thermoanaerobacter kivui (bacteria) | ||||||||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.4 Å cryo EM / Resolution: 3.4 Å | ||||||||||||||||||
 Authors Authors | Dietrich HM / Righetto RD / Kumar A / Wietrzynski W / Schuller SK / Trischler R / Wagner J / Schwarz FM / Engel BD / Mueller V / Schuller JM | ||||||||||||||||||
| Funding support |  Germany, European Union, 5 items Germany, European Union, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Membrane-anchored HDCR nanowires drive hydrogen-powered CO fixation. Authors: Helge M Dietrich / Ricardo D Righetto / Anuj Kumar / Wojciech Wietrzynski / Raphael Trischler / Sandra K Schuller / Jonathan Wagner / Fabian M Schwarz / Benjamin D Engel / Volker Müller / Jan M Schuller /   Abstract: Filamentous enzymes have been found in all domains of life, but the advantage of filamentation is often elusive. Some anaerobic, autotrophic bacteria have an unusual filamentous enzyme for CO ...Filamentous enzymes have been found in all domains of life, but the advantage of filamentation is often elusive. Some anaerobic, autotrophic bacteria have an unusual filamentous enzyme for CO fixation-hydrogen-dependent CO reductase (HDCR)-which directly converts H and CO into formic acid. HDCR reduces CO with a higher activity than any other known biological or chemical catalyst, and it has therefore gained considerable interest in two areas of global relevance: hydrogen storage and combating climate change by capturing atmospheric CO. However, the mechanistic basis of the high catalytic turnover rate of HDCR has remained unknown. Here we use cryo-electron microscopy to reveal the structure of a short HDCR filament from the acetogenic bacterium Thermoanaerobacter kivui. The minimum repeating unit is a hexamer that consists of a formate dehydrogenase (FdhF) and two hydrogenases (HydA2) bound around a central core of hydrogenase Fe-S subunits, one HycB3 and two HycB4. These small bacterial polyferredoxin-like proteins oligomerize through their C-terminal helices to form the backbone of the filament. By combining structure-directed mutagenesis with enzymatic analysis, we show that filamentation and rapid electron transfer through the filament enhance the activity of HDCR. To investigate the structure of HDCR in situ, we imaged T. kivui cells with cryo-electron tomography and found that HDCR filaments bundle into large ring-shaped superstructures attached to the plasma membrane. This supramolecular organization may further enhance the stability and connectivity of HDCR to form a specialized metabolic subcompartment within the cell. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14169.map.gz emd_14169.map.gz | 157.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14169-v30.xml emd-14169-v30.xml emd-14169.xml emd-14169.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_14169.png emd_14169.png | 108.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14169 http://ftp.pdbj.org/pub/emdb/structures/EMD-14169 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14169 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14169 | HTTPS FTP |
-Related structure data
| Related structure data |  7qv7MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14169.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14169.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3.4 Angstrom Cryo-EM structure of HDCR complex. The data was processed using cryosparc software. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Hydrogen-dependent CO2 reductase (HDCR) complex with iron-sulphur...
| Entire | Name: Hydrogen-dependent CO2 reductase (HDCR) complex with iron-sulphur clusters and the H-cluster. |
|---|---|
| Components |
|
-Supramolecule #1: Hydrogen-dependent CO2 reductase (HDCR) complex with iron-sulphur...
| Supramolecule | Name: Hydrogen-dependent CO2 reductase (HDCR) complex with iron-sulphur clusters and the H-cluster. type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: A single functional unit of HDCR complex consists of 4 subunits - FdhF, HydA2, HycB3, and HycB4(1) |
|---|---|
| Source (natural) | Organism:    Thermoanaerobacter kivui LKT-1 (bacteria) Thermoanaerobacter kivui LKT-1 (bacteria) |
-Macromolecule #1: Hydrogen dependent carbon dioxide reductase subunit HycB3
| Macromolecule | Name: Hydrogen dependent carbon dioxide reductase subunit HycB3 type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number:  Oxidoreductases Oxidoreductases |
|---|---|
| Source (natural) | Organism:    Thermoanaerobacter kivui (bacteria) Thermoanaerobacter kivui (bacteria) |
| Molecular weight | Theoretical: 20.520934 KDa |
| Sequence | String: MPNRFVIADP KRCLGCYTCI AACAFVHEEQ GLQPFPRLYL TYTSEGIMPI QCRHCEDAPC AEVCPVEAIK KEGNAIIIDE KACIGCKTC LLACSFGAID FSVQDSLEQS IFKDIKENLM QDQKTQQRIV AVKCDLCNFR EEGPACVQFC PTKALKLVDG D EINKMVKN KRTVNVESLL SVYGTK |
-Macromolecule #2: Hydrogen dependent carbon dioxide reductase subunit HycB4
| Macromolecule | Name: Hydrogen dependent carbon dioxide reductase subunit HycB4 type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO / EC number:  Oxidoreductases Oxidoreductases |
|---|---|
| Source (natural) | Organism:    Thermoanaerobacter kivui (bacteria) Thermoanaerobacter kivui (bacteria) |
| Molecular weight | Theoretical: 23.05218 KDa |
| Sequence | String: MYQKVNCYSI LFLKGVDKMK TQLNPFVVAN PAKCIGCKAC EVACFAVHNR NNHVGATVGT VSIPVIPRLH LIKTEHGTMP IQCRHCEDA PCANVCTVGA IKREGNAIVV DEKLCIGCKS CLLACPFGAI ELLPQYEDGR EVFQINLKEE SESGLVQEPR I IAYKCDLC ...String: MYQKVNCYSI LFLKGVDKMK TQLNPFVVAN PAKCIGCKAC EVACFAVHNR NNHVGATVGT VSIPVIPRLH LIKTEHGTMP IQCRHCEDA PCANVCTVGA IKREGNAIVV DEKLCIGCKS CLLACPFGAI ELLPQYEDGR EVFQINLKEE SESGLVQEPR I IAYKCDLC NDLGEPACVK ACPENALTLV MPTEMKKARN KEAALSFLRV VR |
-Macromolecule #3: Hydrogen dependent carbon dioxide reductase subunit HydA2
| Macromolecule | Name: Hydrogen dependent carbon dioxide reductase subunit HydA2 type: protein_or_peptide / ID: 3 / Number of copies: 6 / Enantiomer: LEVO / EC number:  ferredoxin hydrogenase ferredoxin hydrogenase |
|---|---|
| Source (natural) | Organism:    Thermoanaerobacter kivui (bacteria) Thermoanaerobacter kivui (bacteria) |
| Molecular weight | Theoretical: 51.43027 KDa |
| Sequence | String: MSANKAIINI DQELCTGCRR CAEVCPVDAI EGEKGKPQKI NTEVCVMCGQ CVQKCSSYAS YFDESITPRN VKLQERGMLD SVKEPLFAA YNLGYARQVK EALENPQLFK VVQCAPAIRV SIAEEFGLDL GDLTPGKLVA ALRRLNFDRV YDTNFGADLT I IEEANELV ...String: MSANKAIINI DQELCTGCRR CAEVCPVDAI EGEKGKPQKI NTEVCVMCGQ CVQKCSSYAS YFDESITPRN VKLQERGMLD SVKEPLFAA YNLGYARQVK EALENPQLFK VVQCAPAIRV SIAEEFGLDL GDLTPGKLVA ALRRLNFDRV YDTNFGADLT I IEEANELV KRIKEGKDLP MFTSCCPAWV KFAEQTYPEL LKHISTCKSP QQMTGAIIKT YGAKINNVDP AKIFSVSVMP CT CKSYESD RPEMRSSGYK DVDLVITTRE LAHLMKDKGI DFATLPDEEF DSPLGNYTGA ATIFGNTGGV MEAALRTAYE LIT KKPIPN IDIEFVRGGE GIRTATVQVG ELELKIAVVS GLKNVIPILE DIKKNKCDLH FVEVMTCPEG CISGGGQPKL LLEE YREVA YKKRKEALYK HDAELELRKS HENPAIKKLY EEFLGEPLGK QSHHLLHTKY TPRKKV |
-Macromolecule #4: Hydrogen dependent carbon dioxide reductase subunit FdhF
| Macromolecule | Name: Hydrogen dependent carbon dioxide reductase subunit FdhF type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO / EC number:  formate dehydrogenase formate dehydrogenase |
|---|---|
| Source (natural) | Organism:    Thermoanaerobacter kivui (bacteria) Thermoanaerobacter kivui (bacteria) |
| Molecular weight | Theoretical: 82.505711 KDa |
| Sequence | String: MKDGKQEKVL TTCPYCGTGC GLYLKVENEK IVGVEPDKLH PVNQGELCIK GYYGYKYVHD PRRLTSPLIK KNGKFVPVSW DEALNFIAN GLKKIKSEYG SDAFAMFCSA RATNEDNYAA QKFARAVIGI NNVDHCARLC HAPTVAGLAM TLGSGAMTNS I PEISTYSD ...String: MKDGKQEKVL TTCPYCGTGC GLYLKVENEK IVGVEPDKLH PVNQGELCIK GYYGYKYVHD PRRLTSPLIK KNGKFVPVSW DEALNFIAN GLKKIKSEYG SDAFAMFCSA RATNEDNYAA QKFARAVIGI NNVDHCARLC HAPTVAGLAM TLGSGAMTNS I PEISTYSD VIFIIGSNTA ECHPLIAAHV IKAKERGAKL IVADPRMNAM VHKADIWLRV PSGYNIPLIN GMIHIIIKEG LV KTDFVKN HAVGFEEMAK AVEKYTPEYV EELTGIPKKD LIKAARFYGQ AQAAAILYSM GVTQFSHGTG NVVSLANLAV ITG NLGRPG AGICPLRGQN NVQGACDVGA LPNVLPGYLD VTKEQNRERF EKVWGVKLPS NIGLRVTEVP DAILNKRVRA LYIF GENPI MSDPDSDHLR HALEHLDLLI VQDIFLTETA RLAHVVLPAA CWAEKDGTFT NTERRVQRVR KAVEAPGEAK PDWWI FSQI AERMGYTGMQ YNNVQEIWDE VRKIVPEKFG GISYARLEKE KGLAWPCPTE DHTGTPILYL GGKFATPSGK AQMYPV IFY PNTCICDEGA EKQDFNHVIV GSIAELPDEE YPFTLTTGRR VYHYHTATMT RKSPVIDQIA PQELVEINPQ DATRLGI ND GDFLRVSTRR GYVATRAWVT ERVPKGTIFM TFHYWEACCN ELTNTASDAI CCIPEFKVAA AKVEKISQVE AQAILKEK I EKYQVELEKD VANMLAKEKG GK |
-Macromolecule #5: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 5 / Number of copies: 52 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Macromolecule #6: dicarbonyl[bis(cyanide-kappaC)]-mu-(iminodimethanethiolatato-1kap...
| Macromolecule | Name: dicarbonyl[bis(cyanide-kappaC)]-mu-(iminodimethanethiolatato-1kappaS:2kappaS)-mu-(oxomethylidene)diiron(2+) type: ligand / ID: 6 / Number of copies: 6 / Formula: 402 |
|---|---|
| Molecular weight | Theoretical: 354.953 Da |
| Chemical component information | 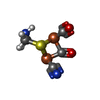 ChemComp-402: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 5.5 µm / Nominal defocus min: 0.5 µm Bright-field microscopy / Nominal defocus max: 5.5 µm / Nominal defocus min: 0.5 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 52.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Initial angle assignment | Type: OTHER |
|---|---|
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 719937 |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




















