[English] 日本語
 Yorodumi
Yorodumi- EMDB-14049: Cryo-EM structure of ABC transporter STE6-2p from Pichia pastoris... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of ABC transporter STE6-2p from Pichia pastoris with Verapamil at 3.2 A resolution | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Komagataella phaffii CBS 7435 (fungus) Komagataella phaffii CBS 7435 (fungus) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.2 Å cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Schleker ESM / Reinhart C | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Structural and functional investigation of ABC transporter STE6-2p from reveals unexpected interaction with sterol molecules. Authors: E Sabine M Schleker / Sabine Buschmann / Hao Xie / Sonja Welsch / Hartmut Michel / Christoph Reinhart /  Abstract: Adenosine triphosphate (ATP)-binding cassette (ABC) transporters are multidomain transmembrane proteins, which facilitate the transport of various substances across cell membranes using energy ...Adenosine triphosphate (ATP)-binding cassette (ABC) transporters are multidomain transmembrane proteins, which facilitate the transport of various substances across cell membranes using energy derived from ATP hydrolysis. They are important drug targets since they mediate decreased drug susceptibility during pharmacological treatments. For the methylotrophic yeast , a model organism that is a widely used host for protein expression, the role and function of its ABC transporters is unexplored. In this work, we investigated the ABC-B transporter STE6-2p. Functional investigations revealed that STE6-2p is capable of transporting rhodamines in vivo and is active in the presence of verapamil and triazoles in vitro. A phylogenetic analysis displays homology among multidrug resistance (MDR) transporters from pathogenic fungi to human ABC-B transporters. Further, we present high-resolution single-particle electron cryomicroscopy structures of an ABC transporter from in the apo conformation (3.1 Å) and in complex with verapamil and adenylyl imidodiphosphate (AMP-PNP) (3.2 Å). An unknown density between transmembrane helices 4, 5, and 6 in both structures suggests the presence of a sterol-binding site of unknown function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14049.map.gz emd_14049.map.gz | 57.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14049-v30.xml emd-14049-v30.xml emd-14049.xml emd-14049.xml | 16.6 KB 16.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14049_fsc.xml emd_14049_fsc.xml | 11.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_14049.png emd_14049.png | 91.6 KB | ||
| Masks |  emd_14049_msk_1.map emd_14049_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Others |  emd_14049_half_map_1.map.gz emd_14049_half_map_1.map.gz emd_14049_half_map_2.map.gz emd_14049_half_map_2.map.gz | 71.2 MB 71.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14049 http://ftp.pdbj.org/pub/emdb/structures/EMD-14049 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14049 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14049 | HTTPS FTP |
-Related structure data
| Related structure data |  7qkrMC  7qksC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14049.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14049.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.837 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14049_msk_1.map emd_14049_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_14049_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_14049_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : STE6-2p
| Entire | Name: STE6-2p |
|---|---|
| Components |
|
-Supramolecule #1: STE6-2p
| Supramolecule | Name: STE6-2p / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Komagataella phaffii CBS 7435 (fungus) / Strain: X-33 Komagataella phaffii CBS 7435 (fungus) / Strain: X-33 |
| Recombinant expression | Organism:  Komagataella phaffii CBS 7435 (fungus) / Recombinant strain: X-33 Komagataella phaffii CBS 7435 (fungus) / Recombinant strain: X-33 |
-Macromolecule #1: Plasma membrane ATP-binding cassette transporter required for the...
| Macromolecule | Name: Plasma membrane ATP-binding cassette transporter required for the export of a-factor type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Komagataella phaffii CBS 7435 (fungus) Komagataella phaffii CBS 7435 (fungus)Strain: ATCC 76273 / CBS 7435 / CECT 11047 / NRRL Y-11430 / Wegner 21-1 |
| Molecular weight | Theoretical: 144.974266 KDa |
| Recombinant expression | Organism:  Komagataella phaffii CBS 7435 (fungus) Komagataella phaffii CBS 7435 (fungus) |
| Sequence | String: MVKNYDYKDD DDKGLSFFKR KKYSDKESLS SPETLEEKPN DELDPRVTEI LERQIKADSY GASLVDLYGM LQGWEYCLAV AAYICSIVA GAALPLMTLI FGDMAQQFTD YSSGLHSNNQ FVDKIDENAL YFVYLGVGLL VFNYFATLLH IVVSEIIASR V REKFIWSI ...String: MVKNYDYKDD DDKGLSFFKR KKYSDKESLS SPETLEEKPN DELDPRVTEI LERQIKADSY GASLVDLYGM LQGWEYCLAV AAYICSIVA GAALPLMTLI FGDMAQQFTD YSSGLHSNNQ FVDKIDENAL YFVYLGVGLL VFNYFATLLH IVVSEIIASR V REKFIWSI LHQNMAYLDS LGSGEITSSI TSDSQLIQQG VSEKIGLAAQ SIATVVSALT VAFVIYWKLA LVLLSVMVAL IL SSTPTIL MLMQAYTDSI ASYGKASSVA EEAFAAIKTA TAFGAHEFQL QKYDEFILES KGYGKKKAIS LALMMGSIWF IVF ATYALA FWQGSRFMVS DNSGIGKILT ACMAMLFGSL IIGNATISLK FVMVGLSAAS KLFAMINREP YFDSASDAGE KINE FDGSI SFRNVTTRYP SRPDITVLSD FTLDIKPGQT IALVGESGSG KSTVIALLER FYEYLDGEIL LDGVDLKSLN IKWVR QQMA LVQQEPVLFA ASIYENVCYG LVGSKYENVT EKVKRELVEK ACKDANAWEF ISQMSNGLDT EVGERGLSLS GGQKQR IAI ARAVISEPKI LLLDEATSAL DTRSEGIVQD ALNRLSETRT TIVIAHRLST IQNADLIVVL SKGKIVETGS HKELLKK KG KYHQLVQIQN IRTKINNSGP QAPISLSNSS DLDSVSHKID RVESLIYERA AADTIDESPV KKQSIPQLFL MLLQINKG D YYLLIPCLFL ALIAGMGFPS FALLAGRVIE AFQVTGPQDF PHMRSLINKY TGFLFMIGCV LLIVYLFLTS FMVLSSESL VYKMRYRCFK QYLRQDMSFF DRPENKVGTL VTTLAKDPQD IEGLSGGTAA QLAVSVVIVV AGIILAVAVN WRLGLVCTAT VPILLGCGF FSVYLLMVFE ERILKDYQES ASYACEQVSA LKTVVSLTRE VGIYEKYSNS IKDQVKRSAR SVSRTTLLYA L IQGMNPWV FALGFWYGSR LLLEGRATNR EFFTVLMAIL FGCQSAGEFF SYAPGMGKAK QAAINIRQVL DTRPKSIDIE SE DGLKIDR LNLKGGIELR DVTFRYPTRP EVPVLTDLNL IIKPGQYVGL VGASGCGKST TVGLIERFYD PESGQVLLDG VDI RDLHLR TYREVLALVQ QEPVLFSGSI RDNIMVGSIS DGADDGSEED MIKACKDANI YDFISSLPEG FDTLCGNKGT MLSG GQKQR VAIARALIRN PRVLLLDEAT SALDSESEMV VQDAIDKASK GRTTITIAHR LSTVQNCDVI YVFDAGRIVE SGKHD ELLQ LRGKYYDLVQ LQGLNAHHHH HHHHHH |
-Macromolecule #2: Dexverapamil
| Macromolecule | Name: Dexverapamil / type: ligand / ID: 2 / Number of copies: 1 / Formula: I6H |
|---|---|
| Molecular weight | Theoretical: 454.602 Da |
| Chemical component information | 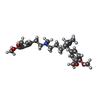 ChemComp-I6H: |
-Macromolecule #3: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER / type: ligand / ID: 3 / Number of copies: 1 / Formula: ANP |
|---|---|
| Molecular weight | Theoretical: 506.196 Da |
| Chemical component information |  ChemComp-ANP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]sp...
| Macromolecule | Name: (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en type: ligand / ID: 5 / Number of copies: 1 / Formula: 9Z9 |
|---|---|
| Molecular weight | Theoretical: 544.805 Da |
| Chemical component information |  ChemComp-9Z9: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 0.0021000000000000003 µm / Nominal defocus min: 0.0011 µm Bright-field microscopy / Nominal defocus max: 0.0021000000000000003 µm / Nominal defocus min: 0.0011 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)













































