+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Jumbo Phage phi-Kp24 empty capsid | |||||||||
 Map data Map data | Klebsiella phage phi-kp24 empty capsid | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Jumbo phage /  Klebsiella pneumoniae / Klebsiella pneumoniae /  Capsid / Capsid /  VIRUS VIRUS | |||||||||
| Function / homology | Major head protein Function and homology information Function and homology information | |||||||||
| Biological species |  Klebsiella phage vB_KpM_FBKp24 (virus) Klebsiella phage vB_KpM_FBKp24 (virus) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.1 Å cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Ouyang R / Briegel A | |||||||||
| Funding support |  Netherlands, 1 items Netherlands, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: High-resolution reconstruction of a Jumbo-bacteriophage infecting capsulated bacteria using hyperbranched tail fibers. Authors: Ruochen Ouyang / Ana Rita Costa / C Keith Cassidy / Aleksandra Otwinowska / Vera C J Williams / Agnieszka Latka / Phill J Stansfeld / Zuzanna Drulis-Kawa / Yves Briers / Daniël M Pelt / ...Authors: Ruochen Ouyang / Ana Rita Costa / C Keith Cassidy / Aleksandra Otwinowska / Vera C J Williams / Agnieszka Latka / Phill J Stansfeld / Zuzanna Drulis-Kawa / Yves Briers / Daniël M Pelt / Stan J J Brouns / Ariane Briegel /      Abstract: The Klebsiella jumbo myophage ϕKp24 displays an unusually complex arrangement of tail fibers interacting with a host cell. In this study, we combine cryo-electron microscopy methods, protein ...The Klebsiella jumbo myophage ϕKp24 displays an unusually complex arrangement of tail fibers interacting with a host cell. In this study, we combine cryo-electron microscopy methods, protein structure prediction methods, molecular simulations, microbiological and machine learning approaches to explore the capsid, tail, and tail fibers of ϕKp24. We determine the structure of the capsid and tail at 4.1 Å and 3.0 Å resolution. We observe the tail fibers are branched and rearranged dramatically upon cell surface attachment. This complex configuration involves fourteen putative tail fibers with depolymerase activity that provide ϕKp24 with the ability to infect a broad panel of capsular polysaccharide (CPS) types of Klebsiella pneumoniae. Our study provides structural and functional insight into how ϕKp24 adapts to the variable surfaces of capsulated bacterial pathogens, which is useful for the development of phage therapy approaches against pan-drug resistant K. pneumoniae strains. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13862.map.gz emd_13862.map.gz | 216.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13862-v30.xml emd-13862-v30.xml emd-13862.xml emd-13862.xml | 12.3 KB 12.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13862_fsc.xml emd_13862_fsc.xml | 28 KB | Display |  FSC data file FSC data file |
| Images |  emd_13862.png emd_13862.png | 251.7 KB | ||
| Filedesc metadata |  emd-13862.cif.gz emd-13862.cif.gz | 3.8 KB | ||
| Others |  emd_13862_half_map_1.map.gz emd_13862_half_map_1.map.gz emd_13862_half_map_2.map.gz emd_13862_half_map_2.map.gz | 1.5 GB 1.5 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13862 http://ftp.pdbj.org/pub/emdb/structures/EMD-13862 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13862 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13862 | HTTPS FTP |
-Related structure data
| Related structure data |  8bflMC  8bfpMC  8au1C  8bfkC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13862.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13862.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Klebsiella phage phi-kp24 empty capsid | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.055 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: phage phi-kp24 empty capsid half 1
| File | emd_13862_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | phage phi-kp24 empty capsid half 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
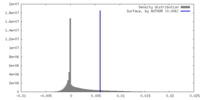
| Density Histograms |
-Half map: phage phi-kp24 empty capsid half 2
| File | emd_13862_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | phage phi-kp24 empty capsid half 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
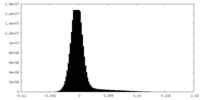
| Density Histograms |
- Sample components
Sample components
-Entire : Phage capisd
| Entire | Name: Phage capisd |
|---|---|
| Components |
|
-Supramolecule #1: Phage capisd
| Supramolecule | Name: Phage capisd / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Klebsiella phage vB_KpM_FBKp24 (virus) Klebsiella phage vB_KpM_FBKp24 (virus) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 64000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 64000 |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average exposure time: 0.053 sec. / Average electron dose: 30.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)