[English] 日本語
 Yorodumi
Yorodumi- EMDB-13829: Cryo-EM structure of Mycobacterium tuberculosis RNA polymerase ho... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Mycobacterium tuberculosis RNA polymerase holoenzyme dimer comprising sigma factor SigB | |||||||||
 Map data Map data | primary map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationRNA polymerase core enzyme binding / Antimicrobial action and antimicrobial resistance in Mtb /  sigma factor activity / peptidoglycan-based cell wall / sigma factor activity / peptidoglycan-based cell wall /  DNA-directed RNA polymerase complex / DNA-templated transcription initiation / DNA-directed RNA polymerase complex / DNA-templated transcription initiation /  ribonucleoside binding / DNA-directed 5'-3' RNA polymerase activity / ribonucleoside binding / DNA-directed 5'-3' RNA polymerase activity /  DNA-directed RNA polymerase / response to heat ...RNA polymerase core enzyme binding / Antimicrobial action and antimicrobial resistance in Mtb / DNA-directed RNA polymerase / response to heat ...RNA polymerase core enzyme binding / Antimicrobial action and antimicrobial resistance in Mtb /  sigma factor activity / peptidoglycan-based cell wall / sigma factor activity / peptidoglycan-based cell wall /  DNA-directed RNA polymerase complex / DNA-templated transcription initiation / DNA-directed RNA polymerase complex / DNA-templated transcription initiation /  ribonucleoside binding / DNA-directed 5'-3' RNA polymerase activity / ribonucleoside binding / DNA-directed 5'-3' RNA polymerase activity /  DNA-directed RNA polymerase / response to heat / response to hypoxia / DNA-directed RNA polymerase / response to heat / response to hypoxia /  protein dimerization activity / response to xenobiotic stimulus / response to antibiotic / DNA-templated transcription / positive regulation of DNA-templated transcription / magnesium ion binding / protein dimerization activity / response to xenobiotic stimulus / response to antibiotic / DNA-templated transcription / positive regulation of DNA-templated transcription / magnesium ion binding /  DNA binding / zinc ion binding / DNA binding / zinc ion binding /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.36 Å cryo EM / Resolution: 4.36 Å | |||||||||
 Authors Authors | Brodolin K | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation | Journal: Nucleic Acids Res / Year: 2014 Title: Mycobacterium RbpA cooperates with the stress-response σB subunit of RNA polymerase in promoter DNA unwinding. Authors: Yangbo Hu / Zakia Morichaud / Ayyappasamy Sudalaiyadum Perumal / Françoise Roquet-Baneres / Konstantin Brodolin /  Abstract: RbpA, a transcriptional activator that is essential for Mycobacterium tuberculosis replication and survival during antibiotic treatment, binds to RNA polymerase (RNAP) in the absence of promoter DNA. ...RbpA, a transcriptional activator that is essential for Mycobacterium tuberculosis replication and survival during antibiotic treatment, binds to RNA polymerase (RNAP) in the absence of promoter DNA. It has been hypothesized that RbpA stimulates housekeeping gene expression by promoting assembly of the σ(A) subunit with core RNAP. Here, using a purified in vitro transcription system of M. tuberculosis, we show that RbpA functions in a promoter-dependent manner as a companion of RNAP essential for promoter DNA unwinding and formation of the catalytically active open promoter complex (RPo). Screening for RbpA activity using a full panel of the M. tuberculosis σ subunits demonstrated that RbpA targets σ(A) and stress-response σ(B), but not the alternative σ subunits from the groups 3 and 4. In contrast to σ(A), the σ(B) subunit activity displayed stringent dependency upon RbpA. These results suggest that RbpA-dependent control of RPo formation provides a mechanism for tuning gene expression during the switch between different physiological states, and in the stress response. #1:  Journal: Biorxiv / Year: 2022 Journal: Biorxiv / Year: 2022Title: Structural basis of the mycobacterial stress-response RNA polymerase auto-inhibition via oligomerization Authors: Morichaud Z / Trapani S / Vishwakarma R / Chaloin L / Lionne C / Lai-Kee-Him J / Bron P / Brodolin K | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13829.map.gz emd_13829.map.gz | 327.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13829-v30.xml emd-13829-v30.xml emd-13829.xml emd-13829.xml | 24.3 KB 24.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13829_fsc.xml emd_13829_fsc.xml | 15.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_13829.png emd_13829.png | 118.6 KB | ||
| Masks |  emd_13829_msk_1.map emd_13829_msk_1.map | 347.6 MB |  Mask map Mask map | |
| Others |  emd_13829_additional_1.map.gz emd_13829_additional_1.map.gz | 154.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13829 http://ftp.pdbj.org/pub/emdb/structures/EMD-13829 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13829 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13829 | HTTPS FTP |
-Related structure data
| Related structure data |  7q59MC  7q4uC  7z8qC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13829.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13829.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | primary map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
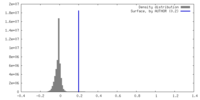
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_13829_msk_1.map emd_13829_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: raw map
| File | emd_13829_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | raw map | ||||||||||||
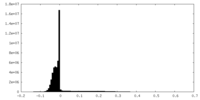
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : RNA polymerase holoenzyme with sigma factor sigB
| Entire | Name: RNA polymerase holoenzyme with sigma factor sigB |
|---|---|
| Components |
|
-Supramolecule #1: RNA polymerase holoenzyme with sigma factor sigB
| Supramolecule | Name: RNA polymerase holoenzyme with sigma factor sigB / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#5 Details: RNA polymerase holoenzyme assembled from individually expressed RNA polymerase core and sigma factor sigB |
|---|---|
| Source (natural) | Organism:   Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) |
| Molecular weight | Theoretical: 800 KDa |
-Macromolecule #1: DNA-directed RNA polymerase subunit alpha
| Macromolecule | Name: DNA-directed RNA polymerase subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number:  DNA-directed RNA polymerase DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:   Mycobacterium tuberculosis H37Rv (bacteria) / Strain: ATCC 25618 / H37Rv Mycobacterium tuberculosis H37Rv (bacteria) / Strain: ATCC 25618 / H37Rv |
| Molecular weight | Theoretical: 37.745328 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: MLISQRPTLS EDVLTDNRSQ FVIEPLEPGF GYTLGNSLRR TLLSSIPGAA VTSIRIDGVL HEFTTVPGVK EDVTEIILNL KSLVVSSEE DEPVTMYLRK QGPGEVTAGD IVPPAGVTVH NPGMHIATLN DKGKLEVELV VERGRGYVPA VQNRASGAEI G RIPVDSIY ...String: MLISQRPTLS EDVLTDNRSQ FVIEPLEPGF GYTLGNSLRR TLLSSIPGAA VTSIRIDGVL HEFTTVPGVK EDVTEIILNL KSLVVSSEE DEPVTMYLRK QGPGEVTAGD IVPPAGVTVH NPGMHIATLN DKGKLEVELV VERGRGYVPA VQNRASGAEI G RIPVDSIY SPVLKVTYKV DATRVEQRTD FDKLILDVET KNSISPRDAL ASAGKTLVEL FGLARELNVE AEGIEIGPSP AE ADHIASF ALPIDDLDLT VRSYNCLKRE GVHTVGELVA RTESDLLDIR NFGQKSIDEV KIKLHQLGLS LKDSPPSFDP SEV AGYDVA TGTWSTEGAY DEQDYAETEQ L |
-Macromolecule #2: DNA-directed RNA polymerase subunit beta
| Macromolecule | Name: DNA-directed RNA polymerase subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO / EC number:  DNA-directed RNA polymerase DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:   Mycobacterium tuberculosis H37Rv (bacteria) / Strain: ATCC 25618 / H37Rv Mycobacterium tuberculosis H37Rv (bacteria) / Strain: ATCC 25618 / H37Rv |
| Molecular weight | Theoretical: 129.602344 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: MVLADSRQSK TAASPSPSRP QSSSNNSVPG APNRVSFAKL REPLEVPGLL DVQTDSFEWL IGSPRWRESA AERGDVNPVG GLEEVLYEL SPIEDFSGSM SLSFSDPRFD DVKAPVDECK DKDMTYAAPL FVTAEFINNN TGEIKSQTVF MGDFPMMTEK G TFIINGTE ...String: MVLADSRQSK TAASPSPSRP QSSSNNSVPG APNRVSFAKL REPLEVPGLL DVQTDSFEWL IGSPRWRESA AERGDVNPVG GLEEVLYEL SPIEDFSGSM SLSFSDPRFD DVKAPVDECK DKDMTYAAPL FVTAEFINNN TGEIKSQTVF MGDFPMMTEK G TFIINGTE RVVVSQLVRS PGVYFDETID KSTDKTLHSV KVIPSRGAWL EFDVDKRDTV GVRIDRKRRQ PVTVLLKALG WT SEQIVER FGFSEIMRST LEKDNTVGTD EALLDIYRKL RPGEPPTKES AQTLLENLFF KEKRYDLARV GRYKVNKKLG LHV GEPITS STLTEEDVVA TIEYLVRLHE GQTTMTVPGG VEVPVETDDI DHFGNRRLRT VGELIQNQIR VGMSRMERVV RERM TTQDV EAITPQTLIN IRPVVAAIKE FFGTSQLSQF MDQNNPLSGL THKRRLSALG PGGLSRERAG LEVRDVHPSH YGRMC PIET PEGPNIGLIG SLSVYARVNP FGFIETPYRK VVDGVVSDEI VYLTADEEDR HVVAQANSPI DADGRFVEPR VLVRRK AGE VEYVPSSEVD YMDVSPRQMV SVATAMIPFL EHDDANRALM GANMQRQAVP LVRSEAPLVG TGMELRAAID AGDVVVA EE SGVIEEVSAD YITVMHDNGT RRTYRMRKFA RSNHGTCANQ CPIVDAGDRV EAGQVIADGP CTDDGEMALG KNLLVAIM P WEGHNYEDAI ILSNRLVEED VLTSIHIEEH EIDARDTKLG AEEITRDIPN ISDEVLADLD ERGIVRIGAE VRDGDILVG KVTPKGETEL TPEERLLRAI FGEKAREVRD TSLKVPHGES GKVIGIRVFS REDEDELPAG VNELVRVYVA QKRKISDGDK LAGRHGNKG VIGKILPVED MPFLADGTPV DIILNTHGVP RRMNIGQILE THLGWCAHSG WKVDAAKGVP DWAARLPDEL L EAQPNAIV STPVFDGAQE AELQGLLSCT LPNRDGDVLV DADGKAMLFD GRSGEPFPYP VTVGYMYIMK LHHLVDDKIH AR STGPYSM ITQQPLGGKA QFGGQRFGEM ECWAMQAYGA AYTLQELLTI KSDDTVGRVK VYEAIVKGEN IPEPGIPESF KVL LKELQS LCLNVEVLSS DGAAIELREG EDEDLERAAA NLGINLSRNE SASVEDLA |
-Macromolecule #3: DNA-directed RNA polymerase subunit beta'
| Macromolecule | Name: DNA-directed RNA polymerase subunit beta' / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO / EC number:  DNA-directed RNA polymerase DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:   Mycobacterium tuberculosis H37Rv (bacteria) / Strain: ATCC 25618 / H37Rv Mycobacterium tuberculosis H37Rv (bacteria) / Strain: ATCC 25618 / H37Rv |
| Molecular weight | Theoretical: 147.438344 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: VNFFDELRIG LATAEDIRQW SYGEVKKPET INYRTLKPEK DGLFCEKIFG PTRDWECYCG KYKRVRFKGI ICERCGVEVT RAKVRRERM GHIELAAPVT HIWYFKGVPS RLGYLLDLAP KDLEKIIYFA AYVITSVDEE MRHNELSTLE AEMAVERKAV E DQRDGELE ...String: VNFFDELRIG LATAEDIRQW SYGEVKKPET INYRTLKPEK DGLFCEKIFG PTRDWECYCG KYKRVRFKGI ICERCGVEVT RAKVRRERM GHIELAAPVT HIWYFKGVPS RLGYLLDLAP KDLEKIIYFA AYVITSVDEE MRHNELSTLE AEMAVERKAV E DQRDGELE ARAQKLEADL AELEAEGAKA DARRKVRDGG EREMRQIRDR AQRELDRLED IWSTFTKLAP KQLIVDENLY RE LVDRYGE YFTGAMGAES IQKLIENFDI DAEAESLRDV IRNGKGQKKL RALKRLKVVA AFQQSGNSPM GMVLDAVPVI PPE LRPMVQ LDGGRFATSD LNDLYRRVIN RNNRLKRLID LGAPEIIVNN EKRMLQESVD ALFDNGRRGR PVTGPGNRPL KSLS DLLKG KQGRFRQNLL GKRVDYSGRS VIVVGPQLKL HQCGLPKLMA LELFKPFVMK RLVDLNHAQN IKSAKRMVER QRPQV WDVL EEVIAEHPVL LNRAPTLHRL GIQAFEPMLV EGKAIQLHPL VCEAFNADFD GDQMAVHLPL SAEAQAEARI LMLSSN NIL SPASGRPLAM PRLDMVTGLY YLTTEVPGDT GEYQPASGDH PETGVYSSPA EAIMAADRGV LSVRAKIKVR LTQLRPP VE IEAELFGHSG WQPGDAWMAE TTLGRVMFNE LLPLGYPFVN KQMHKKVQAA IINDLAERYP MIVVAQTVDK LKDAGFYW A TRSGVTVSMA DVLVPPRKKE ILDHYEERAD KVEKQFQRGA LNHDERNEAL VEIWKEATDE VGQALREHYP DDNPIITIV DSGATGNFTQ TRTLAGMKGL VTNPKGEFIP RPVKSSFREG LTVLEYFINT HGARKGLADT ALRTADSGYL TRRLVDVSQD VIVREHDCQ TERGIVVELA ERAPDGTLIR DPYIETSAYA RTLGTDAVDE AGNVIVERGQ DLGDPEIDAL LAAGITQVKV R SVLTCATS TGVCATCYGR SMATGKLVDI GEAVGIVAAQ SIGEPGTQLT MRTFHQGGVG EDITGGLPRV QELFEARVPR GK APIADVT GRVRLEDGER FYKITIVPDD GGEEVVYDKI SKRQRLRVFK HEDGSERVLS DGDHVEVGQQ LMEGSADPHE VLR VQGPRE VQIHLVREVQ EVYRAQGVSI HDKHIEVIVR QMLRRVTIID SGSTEFLPGS LIDRAEFEAE NRRVVAEGGE PAAG RPVLM GITKASLATD SWLSAASFQE TTRVLTDAAI NCRSDKLNGL KENVIIGKLI PAGTGINRYR NIAVQPTEEA RAAAY TIPS YEDQYYSPDF GAATGAAVPL DDYGYSDYRH HHHHH |
-Macromolecule #4: DNA-directed RNA polymerase subunit omega
| Macromolecule | Name: DNA-directed RNA polymerase subunit omega / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO / EC number:  DNA-directed RNA polymerase DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:   Mycobacterium tuberculosis H37Rv (bacteria) / Strain: ATCC 25618 / H37Rv Mycobacterium tuberculosis H37Rv (bacteria) / Strain: ATCC 25618 / H37Rv |
| Molecular weight | Theoretical: 11.85114 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: MSISQSDASL AAVPAVDQFD PSSGASGGYD TPLGITNPPI DELLDRVSSK YALVIYAAKR ARQINDYYNQ LGEGILEYVG PLVEPGLQE KPLSIALREI HADLLEHTEG E |
-Macromolecule #5: RNA polymerase sigma factor SigB
| Macromolecule | Name: RNA polymerase sigma factor SigB / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mycobacterium tuberculosis H37Rv (bacteria) / Strain: ATCC 25618 / H37Rv Mycobacterium tuberculosis H37Rv (bacteria) / Strain: ATCC 25618 / H37Rv |
| Molecular weight | Theoretical: 38.572773 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MADAPTRATT SRVDSDLDAQ SPAADLVRVY LNGIGKTALL NAAGEVELAK RIEAGLYAEH LLETRKRLG ENRKRDLAAV VRDGEAARRH LLEANLRLVV SLAKRYTGRG MPLLDLIQEG NLGLIRAMEK FDYTKGFKFS T YATWWIRQ ...String: MGSSHHHHHH SSGLVPRGSH MADAPTRATT SRVDSDLDAQ SPAADLVRVY LNGIGKTALL NAAGEVELAK RIEAGLYAEH LLETRKRLG ENRKRDLAAV VRDGEAARRH LLEANLRLVV SLAKRYTGRG MPLLDLIQEG NLGLIRAMEK FDYTKGFKFS T YATWWIRQ AITRGMADQS RTIRLPVHLV EQVNKLARIK REMHQHLGRE ATDEELAAES GIPIDKINDL LEHSRDPVSL DM PVGSEEE APLGDFIEDA EAMSAENAVI AELLHTDIRS VLATLDEREH QVIRLRFGLD DGQPRTLDQI GKLFGLSRER VRQ IERDVM SKLRHGERAD RLRSYAS |
-Macromolecule #6: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 6 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #7: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 7 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.4 mg/mL |
|---|---|
| Buffer | pH: 7.9 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 7.0 µm / Nominal defocus min: 0.5 µm Bright-field microscopy / Nominal defocus max: 7.0 µm / Nominal defocus min: 0.5 µm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number real images: 3064 / Average electron dose: 49.6 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)