+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11117 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
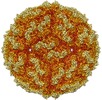
| Title | Human Picobirnavirus D45-CP VLP | |||||||||
 Map data Map data | HPBV delta45 VLP | |||||||||
 Sample Sample | Human picobirnavirus != Human picobirnavirus (strain Human/Thailand/Hy005102/-) Human picobirnavirus
| |||||||||
 Keywords Keywords |  Human Picobirnavirus / Human Picobirnavirus /  CryoEM / assembly / CryoEM / assembly /  capsid protein / capsid protein /  VIRUS LIKE PARTICLE VIRUS LIKE PARTICLE | |||||||||
| Function / homology |  : / : /  : / : /  Picobirnavirus, Capsid protein / T=3 icosahedral viral capsid / Picobirnavirus, Capsid protein / T=3 icosahedral viral capsid /  Capsid protein precursor Capsid protein precursor Function and homology information Function and homology information | |||||||||
| Biological species |   Human picobirnavirus (strain Human/Thailand/Hy005102/-) Human picobirnavirus (strain Human/Thailand/Hy005102/-) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.8 Å cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Ortega-Esteban A / Mata CP | |||||||||
| Funding support |  Spain, 1 items Spain, 1 items
| |||||||||
 Citation Citation |  Journal: J Virol / Year: 2020 Journal: J Virol / Year: 2020Title: Cryo-electron Microscopy Structure, Assembly, and Mechanics Show Morphogenesis and Evolution of Human Picobirnavirus. Authors: Álvaro Ortega-Esteban / Carlos P Mata / María J Rodríguez-Espinosa / Daniel Luque / Nerea Irigoyen / Javier M Rodríguez / Pedro J de Pablo / José R Castón /  Abstract: Despite their diversity, most double-stranded-RNA (dsRNA) viruses share a specialized T=1 capsid built from dimers of a single protein that provides a platform for genome transcription and ...Despite their diversity, most double-stranded-RNA (dsRNA) viruses share a specialized T=1 capsid built from dimers of a single protein that provides a platform for genome transcription and replication. This ubiquitous capsid remains structurally undisturbed throughout the viral cycle, isolating the genome to avoid triggering host defense mechanisms. Human picobirnavirus (hPBV) is a dsRNA virus frequently associated with gastroenteritis, although its pathogenicity is yet undefined. Here, we report the cryo-electron microscopy (cryo-EM) structure of hPBV at 2.6-Å resolution. The capsid protein (CP) is arranged in a single-shelled, ∼380-Å-diameter T=1 capsid with a rough outer surface similar to that of dsRNA mycoviruses. The hPBV capsid is built of 60 quasisymmetric CP dimers (A and B) stabilized by domain swapping, and only the CP-A N-terminal basic region interacts with the packaged nucleic acids. hPBV CP has an α-helical domain with a fold similar to that of fungal partitivirus CP, with many domain insertions in its C-terminal half. In contrast to dsRNA mycoviruses, hPBV has an extracellular life cycle phase like complex reoviruses, which indicates that its own CP probably participates in cell entry. Using an reversible assembly/disassembly system of hPBV, we isolated tetramers as possible assembly intermediates. We used atomic force microscopy to characterize the biophysical properties of hPBV capsids with different cargos (host nucleic acids or proteins) and found that the CP N-terminal segment not only is involved in nucleic acid interaction/packaging but also modulates the mechanical behavior of the capsid in conjunction with the cargo. Despite intensive study, human virus sampling is still sparse, especially for viruses that cause mild or asymptomatic disease. Human picobirnavirus (hPBV) is a double-stranded-RNA virus, broadly dispersed in the human population, but its pathogenicity is uncertain. Here, we report the hPBV structure derived from cryo-electron microscopy (cryo-EM) and reconstruction methods using three capsid protein variants (of different lengths and N-terminal amino acid compositions) that assemble as virus-like particles with distinct properties. The hPBV near-atomic structure reveals a quasisymmetric dimer as the structural subunit and tetramers as possible assembly intermediates that coassemble with nucleic acids. Our structural studies and atomic force microscopy analyses indicate that hPBV capsids are potentially excellent nanocages for gene therapy and targeted drug delivery in humans. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11117.map.gz emd_11117.map.gz | 227.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11117-v30.xml emd-11117-v30.xml emd-11117.xml emd-11117.xml | 12.3 KB 12.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_11117.png emd_11117.png | 337.9 KB | ||
| Filedesc metadata |  emd-11117.cif.gz emd-11117.cif.gz | 5.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11117 http://ftp.pdbj.org/pub/emdb/structures/EMD-11117 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11117 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11117 | HTTPS FTP |
-Related structure data
| Related structure data |  6z8fMC  6z8dC  6z8eC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11117.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11117.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HPBV delta45 VLP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.065 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human picobirnavirus
| Entire | Name:   Human picobirnavirus Human picobirnavirus |
|---|---|
| Components |
|
-Supramolecule #1: Human picobirnavirus (strain Human/Thailand/Hy005102/-)
| Supramolecule | Name: Human picobirnavirus (strain Human/Thailand/Hy005102/-) type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 647332 Sci species name: Human picobirnavirus (strain Human/Thailand/Hy005102/-) Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Virus shell | Shell ID: 1 / Name: Capsid Protein / Diameter: 380.0 Å / T number (triangulation number): 1 |
-Macromolecule #1: Capsid protein precursor
| Macromolecule | Name: Capsid protein precursor / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human picobirnavirus (strain Human/Thailand/Hy005102/-) Human picobirnavirus (strain Human/Thailand/Hy005102/-) |
| Molecular weight | Theoretical: 57.508102 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: ESTRNDVAWY ARYPHILEEA TRLPFAYPIG QYYDTGYSVA SATEWSKYVD TSLTIPGVMC VNFTPTPGES YNKNSPINIA AQNVYTYVR HMNSGHANYE QADLMMYLLA MDSLYIFHSY VRKILAISKL YTPVNKYFPR ALLVALGVDP EDVFANQAQW E YFVNMVAY ...String: ESTRNDVAWY ARYPHILEEA TRLPFAYPIG QYYDTGYSVA SATEWSKYVD TSLTIPGVMC VNFTPTPGES YNKNSPINIA AQNVYTYVR HMNSGHANYE QADLMMYLLA MDSLYIFHSY VRKILAISKL YTPVNKYFPR ALLVALGVDP EDVFANQAQW E YFVNMVAY RAGAFAAPAS MTYYERHAWM SNGLYVDQDV TRAQIYMFKP TMLWKYENLG TTGTKLVPLM MPKAGDNRKL VD FQVLFNN LVSTMLGDED FGIMSGDVFK AFGADGLVKL LAVDSTTMTL PTYDPLILAQ IHSARAVGAP ILETSTLTGF PGR QWQITQ NPDVNNGAII FHPSFGYDGQ DHEELSFRAM CSNMILNLPG EAHSAEMIIE ATRLATMFQV KAVPAGDTSK PVLY LPNGF GTEVVNDYTM ISVDKATPHD LTIHTFFNNI LVPNAKENYV ANLELLNNII QFDWAPQLYL TYGIAQESFG PFAQL NDWT ILTGETLARM HEVCVTSMFD VPQMGFNK UniProtKB:  Capsid protein precursor Capsid protein precursor |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| ||||||
|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM CPC |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 4.04 µm / Calibrated defocus min: 0.306 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 75000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number real images: 1990 / Average electron dose: 1.56 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: Human Picobirnavirus CP model |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 2.1) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 2.1) |
| Final reconstruction | Applied symmetry - Point group: I (icosahedral ) / Resolution.type: BY AUTHOR / Resolution: 2.8 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 2.1) / Number images used: 51194 ) / Resolution.type: BY AUTHOR / Resolution: 2.8 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 2.1) / Number images used: 51194 |
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-6z8f: |
 Movie
Movie Controller
Controller