[English] 日本語
 Yorodumi
Yorodumi- EMDB-30800: Acidic stable capsid structure of Helicobacter pylori bacteriopha... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30800 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Acidic stable capsid structure of Helicobacter pylori bacteriophage KHP40 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology | Protein of unknown function DUF4043 / Protein of unknown function (DUF4043) / Fn3_like domain-containing protein / Uncharacterized protein Function and homology information Function and homology information | |||||||||
| Biological species |  Helicobacter phage KHP40 (virus) / Helicobacter phage KHP40 (virus) /  Helicobacter phage KHP (virus) Helicobacter phage KHP (virus) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.0 Å cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Kamiya R / Uchiyama J / Matsuzaki S / Murata K / Iwasaki K / Miyazaki N | |||||||||
| Funding support |  Japan, 2 items Japan, 2 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2022 Journal: Structure / Year: 2022Title: Acid-stable capsid structure of Helicobacter pylori bacteriophage KHP30 by single-particle cryoelectron microscopy. Authors: Ryosuke Kamiya / Jumpei Uchiyama / Shigenobu Matsuzaki / Kazuyoshi Murata / Kenji Iwasaki / Naoyuki Miyazaki /  Abstract: The acid-stable capsid structures of Helicobacter pylori phages KHP30 and KHP40 are solved at 2.7 and 3.0 Å resolutions by cryoelectron microscopy, respectively. The capsids have icosahedral T = 9 ...The acid-stable capsid structures of Helicobacter pylori phages KHP30 and KHP40 are solved at 2.7 and 3.0 Å resolutions by cryoelectron microscopy, respectively. The capsids have icosahedral T = 9 symmetry and consist of each 540 copies of 2 structural proteins, a major capsid protein, and a cement protein. The major capsid proteins form 12 pentagonal capsomeres occupying icosahedral vertexes and 80 hexagonal capsomeres located at icosahedral faces and edges. The major capsid protein has a unique protruding loop extending to the neighboring subunit that stabilizes hexagonal capsomeres. Furthermore, the capsid is decorated with trimeric cement proteins with a jelly roll motif. The cement protein trimer sits on the quasi-three-fold axis formed by three major capsid protein capsomeres, thereby enhancing the particle stability by connecting these capsomeres. Sequence and structure comparisons between the related Helicobacter pylori phages suggest a possible mechanism of phage adaptation to the human gastric environment. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30800.map.gz emd_30800.map.gz | 345.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30800-v30.xml emd-30800-v30.xml emd-30800.xml emd-30800.xml | 10.3 KB 10.3 KB | Display Display |  EMDB header EMDB header |
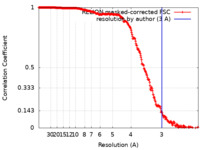
| FSC (resolution estimation) |  emd_30800_fsc.xml emd_30800_fsc.xml | 26.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_30800.png emd_30800.png | 266.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30800 http://ftp.pdbj.org/pub/emdb/structures/EMD-30800 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30800 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30800 | HTTPS FTP |
-Related structure data
| Related structure data |  7douMC  7f2pMC  7dn2C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_30800.map.gz / Format: CCP4 / Size: 1.6 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30800.map.gz / Format: CCP4 / Size: 1.6 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.16 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Helicobacter phage KHP
| Entire | Name:  Helicobacter phage KHP (virus) Helicobacter phage KHP (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Helicobacter phage KHP
| Supramolecule | Name: Helicobacter phage KHP / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 1208236 / Sci species name: Helicobacter phage KHP / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|---|
| Virus shell | Shell ID: 1 / Name: Head / Diameter: 700.0 Å / T number (triangulation number): 9 |
-Macromolecule #1: Cement protein gp16
| Macromolecule | Name: Cement protein gp16 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Helicobacter phage KHP40 (virus) Helicobacter phage KHP40 (virus) |
| Molecular weight | Theoretical: 13.469469 KDa |
| Sequence | String: MKQKVHSVSY LAKAEFEYKN GVYDLVALPT GAEVIKISLE VVGLPTAGHV SVGFKDESKK NYSSILTLPV NETSGVVTKD YTVKSDKIV AAEVKDALAE GSDGRPVKCV LRALYFLPSV IEVEY |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average exposure time: 1.0 sec. / Average electron dose: 20.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller