[English] 日本語
 Yorodumi
Yorodumi- EMDB-20217: Structure of SIVsmm Nef and SMM tetherin bound to the clathrin ad... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20217 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
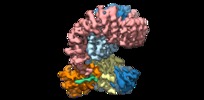
| Title | Structure of SIVsmm Nef and SMM tetherin bound to the clathrin adaptor AP-2 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationAP-type membrane coat adaptor complex / Formation of annular gap junctions / Gap junction degradation / LDL clearance / WNT5A-dependent internalization of FZD2, FZD5 and ROR2 / WNT5A-dependent internalization of FZD2, FZD5 and ROR2 / LDL clearance / WNT5A-dependent internalization of FZD4 / VLDLR internalisation and degradation / Retrograde neurotrophin signalling ...AP-type membrane coat adaptor complex / Formation of annular gap junctions / Gap junction degradation / LDL clearance / WNT5A-dependent internalization of FZD2, FZD5 and ROR2 / WNT5A-dependent internalization of FZD2, FZD5 and ROR2 / LDL clearance / WNT5A-dependent internalization of FZD4 / VLDLR internalisation and degradation / Retrograde neurotrophin signalling / Trafficking of GluR2-containing AMPA receptors / Retrograde neurotrophin signalling / clathrin coat / clathrin adaptor complex / WNT5A-dependent internalization of FZD4 /  extrinsic component of presynaptic endocytic zone membrane / VLDLR internalisation and degradation / cardiac septum development / virus-mediated perturbation of host defense response => GO:0019049 / MHC class II antigen presentation / Recycling pathway of L1 / AP-2 adaptor complex / regulation of vesicle size / postsynaptic neurotransmitter receptor internalization / Recycling pathway of L1 / clathrin coat assembly / Cargo recognition for clathrin-mediated endocytosis / Cargo recognition for clathrin-mediated endocytosis / positive regulation of synaptic vesicle endocytosis / extrinsic component of presynaptic endocytic zone membrane / VLDLR internalisation and degradation / cardiac septum development / virus-mediated perturbation of host defense response => GO:0019049 / MHC class II antigen presentation / Recycling pathway of L1 / AP-2 adaptor complex / regulation of vesicle size / postsynaptic neurotransmitter receptor internalization / Recycling pathway of L1 / clathrin coat assembly / Cargo recognition for clathrin-mediated endocytosis / Cargo recognition for clathrin-mediated endocytosis / positive regulation of synaptic vesicle endocytosis /  Clathrin-mediated endocytosis / clathrin adaptor activity / Clathrin-mediated endocytosis / clathrin adaptor activity /  Clathrin-mediated endocytosis / vesicle budding from membrane / membrane coat / Clathrin-mediated endocytosis / vesicle budding from membrane / membrane coat /  clathrin-dependent endocytosis / MHC class II antigen presentation / neurotransmitter receptor internalization / positive regulation of protein localization to membrane / coronary vasculature development / clathrin-dependent endocytosis / MHC class II antigen presentation / neurotransmitter receptor internalization / positive regulation of protein localization to membrane / coronary vasculature development /  signal sequence binding / negative regulation of protein localization to plasma membrane / aorta development / Neutrophil degranulation / ventricular septum development / low-density lipoprotein particle receptor binding / signal sequence binding / negative regulation of protein localization to plasma membrane / aorta development / Neutrophil degranulation / ventricular septum development / low-density lipoprotein particle receptor binding /  clathrin binding / protein serine/threonine kinase binding / positive regulation of endocytosis / Trafficking of GluR2-containing AMPA receptors / positive regulation of receptor internalization / synaptic vesicle endocytosis / vesicle-mediated transport / clathrin binding / protein serine/threonine kinase binding / positive regulation of endocytosis / Trafficking of GluR2-containing AMPA receptors / positive regulation of receptor internalization / synaptic vesicle endocytosis / vesicle-mediated transport /  phosphatidylinositol binding / phosphatidylinositol binding /  secretory granule / secretory granule /  kidney development / kidney development /  intracellular protein transport / cytoplasmic side of plasma membrane / intracellular protein transport / cytoplasmic side of plasma membrane /  kinase binding / disordered domain specific binding / kinase binding / disordered domain specific binding /  synaptic vesicle / presynapse / synaptic vesicle / presynapse /  heart development / cytoplasmic vesicle / postsynapse / protein-containing complex assembly / defense response to virus / transmembrane transporter binding / membrane => GO:0016020 / protein domain specific binding / intracellular membrane-bounded organelle / glutamatergic synapse / heart development / cytoplasmic vesicle / postsynapse / protein-containing complex assembly / defense response to virus / transmembrane transporter binding / membrane => GO:0016020 / protein domain specific binding / intracellular membrane-bounded organelle / glutamatergic synapse /  lipid binding / lipid binding /  synapse / protein-containing complex binding / GTP binding / synapse / protein-containing complex binding / GTP binding /  protein kinase binding / protein kinase binding /  mitochondrion / mitochondrion /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Rattus norvegicus (Norway rat) / Rattus norvegicus (Norway rat) /   Mus musculus (house mouse) / Mus musculus (house mouse) /   Simian immunodeficiency virus Simian immunodeficiency virus | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.8 Å cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Buffalo CZ / Ren X / Hurley JH | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Host Microbe / Year: 2019 Journal: Cell Host Microbe / Year: 2019Title: Structural Basis for Tetherin Antagonism as a Barrier to Zoonotic Lentiviral Transmission. Authors: Cosmo Z Buffalo / Christina M Stürzel / Elena Heusinger / Dorota Kmiec / Frank Kirchhoff / James H Hurley / Xuefeng Ren /   Abstract: Tetherin is a host defense factor that physically prevents virion release from the plasma membrane. The Nef accessory protein of simian immunodeficiency virus (SIV) engages the clathrin adaptor AP-2 ...Tetherin is a host defense factor that physically prevents virion release from the plasma membrane. The Nef accessory protein of simian immunodeficiency virus (SIV) engages the clathrin adaptor AP-2 to downregulate tetherin via its DIWK motif. As human tetherin lacks DIWK, antagonism of tetherin by Nef is a barrier to simian-human transmission of non-human primate lentiviruses. To determine the molecular basis for tetherin counteraction, we reconstituted the AP-2 complex with a simian tetherin and SIV Nef and determined its structure by cryoelectron microscopy (cryo-EM). Nef refolds the first α-helix of the β2 subunit of AP-2 to a β hairpin, creating a binding site for the DIWK sequence. The tetherin binding site in Nef is distinct from those of most other Nef substrates, including MHC class I, CD3, and CD4 but overlaps with the site for the restriction factor SERINC5. This structure explains the dependence of SIVs on tetherin DIWK and consequent barrier to human transmission. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20217.map.gz emd_20217.map.gz | 1.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20217-v30.xml emd-20217-v30.xml emd-20217.xml emd-20217.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20217_fsc.xml emd_20217_fsc.xml | 9.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_20217.png emd_20217.png | 205.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20217 http://ftp.pdbj.org/pub/emdb/structures/EMD-20217 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20217 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20217 | HTTPS FTP |
-Related structure data
| Related structure data |  6owtMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20217.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20217.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.149 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Structure of SIVsmm Nef and SMM tetherin bound to the clathrin ad...
| Entire | Name: Structure of SIVsmm Nef and SMM tetherin bound to the clathrin adaptor AP-2 complex |
|---|---|
| Components |
|
-Supramolecule #1: Structure of SIVsmm Nef and SMM tetherin bound to the clathrin ad...
| Supramolecule | Name: Structure of SIVsmm Nef and SMM tetherin bound to the clathrin adaptor AP-2 complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Rattus norvegicus (Norway rat) Rattus norvegicus (Norway rat) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Experimental: 203.6 KDa |
-Macromolecule #1: AP-2 complex subunit alpha
| Macromolecule | Name: AP-2 complex subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Rattus norvegicus (Norway rat) Rattus norvegicus (Norway rat) |
| Molecular weight | Theoretical: 104.286391 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MPAVSKGEGM RGLAVFISDI RNCKSKEAEI KRINKELANI RSKFKGDKAL DGYSKKKYVC KLLFIFLLGH DIDFGHMEAV NLLSSNRYT EKQIGYLFIS VLVNSNSELI RLINNAIKND LASRNPTFMG LALHCIANVG SREMAEAFAG EIPKILVAGD T MDSVKQSA ...String: MPAVSKGEGM RGLAVFISDI RNCKSKEAEI KRINKELANI RSKFKGDKAL DGYSKKKYVC KLLFIFLLGH DIDFGHMEAV NLLSSNRYT EKQIGYLFIS VLVNSNSELI RLINNAIKND LASRNPTFMG LALHCIANVG SREMAEAFAG EIPKILVAGD T MDSVKQSA ALCLLRLYRT SPDLVPMGDW TSRVVHLLND QHLGVVTAAT SLITTLAQKN PEEFKTSVSL AVSRLSRIVT SA STDLQDY TYYFVPAPWL SVKLLRLLQC YPPPEDPAVR GRLTECLETI LNKAQEPPKS KKVQHSNAKN AVLFEAISLI IHH DSEPNL LVRACNQLGQ FLQHRETNLR YLALESMCTL ASSEFSHEAV KTHIETVINA LKTERDVSVR QRAVDLLYAM CDRS NAQQI VAEMLSYLET ADYSIREEIV LKVAILAEKY AVDYTWYVDT ILNLIRIAGD YVSEEVWYRV IQIVINRDDV QGYAA KTVF EALQAPACHE NLVKVGGYIL GEFGNLIAGD PRSSPLIQFN LLHSKFHLCS VPTRALLLST YIKFVNLFPE VKATIQ DVL RSDSQLKNAD VELQQRAVEY LRLSTVASTD ILATVLEEMP PFPERESSIL AKLKKKKGPS TVTDLEETKR ERSIDVN GG PEPVPASTSA ASTPSPSADL LGLGAVPPAP TGPPPTSGGG LLVDVFSDSA SAVAPLAPGS EDNFARFVCK NNGVLFEN Q LLQIGLKSEF RQNLGRMFIF YGNKTSTQFL NFTPTLICAD DLQTNLNLQT KPVDPTVDGG AQVQQVVNIE CISDFTEAP VLNIQFRYGG TFQNVSVKLP ITLNKFFQPT EMASQDFFQR WKQLSNPQQE VQNIFKAKHP MDTEITKAKI IGFGSALLEE VDPNPANFV GAGIIHTKTT QIGCLLRLEP NLQAQMYRLT LRTSKDTVSQ RLCELLSEQF |
-Macromolecule #2: AP-2 complex subunit beta
| Macromolecule | Name: AP-2 complex subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Rattus norvegicus (Norway rat) Rattus norvegicus (Norway rat) |
| Molecular weight | Theoretical: 66.953195 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MTDSKYFTTN KKGEIFELKA ELNNEKKEKR KEAVKKVIAA MTVGKDVSSL FPDVVNCMQT DNLELKKLVY LYLMNYAKSQ PDMAIMAVN SFVKDCEDPN PLIRALAVRT MGCIRVDKIT EYLCEPLRKC LKDEDPYVRK TAAVCVAKLH DINAQMVEDQ G FLDSLRDL ...String: MTDSKYFTTN KKGEIFELKA ELNNEKKEKR KEAVKKVIAA MTVGKDVSSL FPDVVNCMQT DNLELKKLVY LYLMNYAKSQ PDMAIMAVN SFVKDCEDPN PLIRALAVRT MGCIRVDKIT EYLCEPLRKC LKDEDPYVRK TAAVCVAKLH DINAQMVEDQ G FLDSLRDL IADSNPMVVA NAVAALSEIS ESHPNSNLLD LNPQNINKLL TALNECTEWG QIFILDCLSN YNPKDDREAQ SI CERVTPR LSHANSAVVL SAVKVLMKFL ELLPKDSDYY NMLLKKLAPP LVTLLSGEPE VQYVALRNIN LIVQKRPEIL KQE IKVFFV KYNDPIYVKL EKLDIMIRLA SQANIAQVLA ELKEYATEVD VDFVRKAVRA IGRCAIKVEQ SAERCVSTLL DLIQ TKVNY VVQEAIVVIR DIFRKYPNKY ESIIATLCEN LDSLDEPDAR AAMIWIVGEY AERIDNADEL LESFLEGFHD ESTQV QLTL LTAIVKLFLK KPSETQELVQ QVLSLATQDS DNPDLRDRGY IYWRLLSTDP VTAKEVVLSE KPLISEETDL IEPTLL DEL ICHIGSLASV YHKPPNAFVE GSHGIHRK |
-Macromolecule #3: Adaptor protein complex AP-2, mu1
| Macromolecule | Name: Adaptor protein complex AP-2, mu1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 16.161563 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MIGGLFIYNH KGEVLISRVY RDDIGRNAVD AFRVNVIHAR QQVRSPVTNI ARTSFFHVKR SNIWLAAVTK QNVNAAMVFE FLYKMCDVM AAYFGKISEE NIKNNFVLIY ELLDEILDFG YPQNSETGAL KTFITQQGIK SQ |
-Macromolecule #4: Tetherin,Protein Nef
| Macromolecule | Name: Tetherin,Protein Nef / type: protein_or_peptide / ID: 4 Details: protein chimera of the cytoplasmic tail of sooty mangabey (smm) linked to the N-terminus of SIV smm tetherin Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Simian immunodeficiency virus Simian immunodeficiency virus |
| Molecular weight | Theoretical: 31.772352 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: ILYDYSRMPM GDIWKEDGDK RSKGSDEASE GSGMGGVTSK KQSKRRQGLR ERLLQARGET DGYSRSRGEL GKGWNLPSAE GQGYSEEQF MNTPWRNPAR EGEKLKYRQQ NMDDVDDDDD ELVGVAVHPK VPLRAMSYKL AIDMSHFIKE KGGLEGIYYS D RRHRILDI ...String: ILYDYSRMPM GDIWKEDGDK RSKGSDEASE GSGMGGVTSK KQSKRRQGLR ERLLQARGET DGYSRSRGEL GKGWNLPSAE GQGYSEEQF MNTPWRNPAR EGEKLKYRQQ NMDDVDDDDD ELVGVAVHPK VPLRAMSYKL AIDMSHFIKE KGGLEGIYYS D RRHRILDI YLEKEEGIIP DWQNYTSGPG VRYPLFFGWL WKLVPVNVSD EAQEDETHCL VHPAQTSQWD DPWGEVLAWK FD PKLAYTY EAFIRYPEEF GNDSGLSKEE VKRRLTAR |
-Macromolecule #5: AP-2 complex subunit sigma
| Macromolecule | Name: AP-2 complex subunit sigma / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Rattus norvegicus (Norway rat) Rattus norvegicus (Norway rat) |
| Molecular weight | Theoretical: 17.011662 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MIRFILIQNR AGKTRLAKWY MQFDDDEKQK LIEEVHAVVT VRDAKHTNFV EFRNFKIIYR RYAGLYFCIC VDVNDNNLAY LEAIHNFVE VLNEYFHCVS ELDLVFNFYK VYTVVDEMFL AGEIRETSQT KVLKQLLMLQ SLE |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: C-flat-2/1 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.04 kPa / Details: 25mA |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 50.16 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller