+Search query
-Structure paper
| Title | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD. |
|---|---|
| Journal, issue, pages | Protein Cell, Vol. 15, Issue 2, Page 121-134, Year 2024 |
| Publish date | Feb 1, 2024 |
 Authors Authors | Hui Sun / Tingting Deng / Yali Zhang / Yanling Lin / Yanan Jiang / Yichao Jiang / Yang Huang / Shuo Song / Lingyan Cui / Tingting Li / Hualong Xiong / Miaolin Lan / Liqin Liu / Yu Li / Qianjiao Fang / Kunyu Yu / Wenling Jiang / Lizhi Zhou / Yuqiong Que / Tianying Zhang / Quan Yuan / Tong Cheng / Zheng Zhang / Hai Yu / Jun Zhang / Wenxin Luo / Shaowei Li / Qingbing Zheng / Ying Gu / Ningshao Xia /  |
| PubMed Abstract | Continual evolution of the severe acute respiratory syndrome coronavirus (SARS-CoV-2) virus has allowed for its gradual evasion of neutralizing antibodies (nAbs) produced in response to natural ...Continual evolution of the severe acute respiratory syndrome coronavirus (SARS-CoV-2) virus has allowed for its gradual evasion of neutralizing antibodies (nAbs) produced in response to natural infection or vaccination. The rapid nature of these changes has incited a need for the development of superior broad nAbs (bnAbs) and/or the rational design of an antibody cocktail that can protect against the mutated virus strain. Here, we report two angiotensin-converting enzyme 2 competing nAbs-8H12 and 3E2-with synergistic neutralization but evaded by some Omicron subvariants. Cryo-electron microscopy reveals the two nAbs synergistic neutralizing virus through a rigorous pairing permitted by rearrangement of the 472-489 loop in the receptor-binding domain to avoid steric clashing. Bispecific antibodies based on these two nAbs tremendously extend the neutralizing breadth and restore neutralization against recent variants including currently dominant XBB.1.5. Together, these findings expand our understanding of the potential strategies for the neutralization of SARS-CoV-2 variants toward the design of broad-acting antibody therapeutics and vaccines. |
 External links External links |  Protein Cell / Protein Cell /  PubMed:37470320 / PubMed:37470320 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.47 - 7.57 Å |
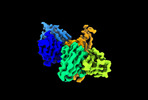
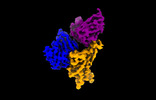
| Structure data | 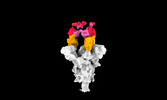 EMDB-35730: Cryo-EM structure of SARS-CoV-2 spike protein in complex with 8H12  EMDB-35731: Cryo-EM structure of SARS-CoV-2 spike protein in complex with 3E2  EMDB-35736: Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 1C4 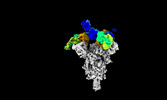 EMDB-35739: Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 3E2 and 1C4 EMDB-35740, PDB-8iv4: EMDB-35741, PDB-8iv5:  EMDB-35743: Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 3E2 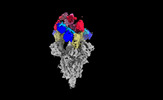 EMDB-35745: cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with double nAbs 8H12 and 3E2 EMDB-35746, PDB-8iv8:  EMDB-35747: cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with double nAbs 8H12 and 3E2 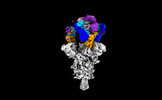 EMDB-35749: Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs XMA01 and 3E2  EMDB-35750: cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs S2E12 and 3E2  EMDB-35751: Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs S2E12 and 3E2 (local refinement)  EMDB-35752: cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and S2X35 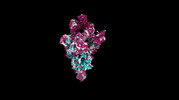 EMDB-35753: cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and VacW-209 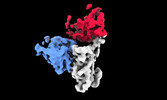 EMDB-35754: cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and S2X35 (local refinement) EMDB-35755, PDB-8iva: |
| Source |
|
 Keywords Keywords |  VIRAL PROTEIN/IMMUNE SYSTEM / VIRAL PROTEIN/IMMUNE SYSTEM /  SARS-CoV-2 / SARS-CoV-2 /  Neutralizing antibody / Neutralizing antibody /  Cryo-EM / Cryo-EM /  VIRAL PROTEIN / VIRAL PROTEIN /  VIRAL PROTEIN-IMMUNE SYSTEM complex VIRAL PROTEIN-IMMUNE SYSTEM complex |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers