+Search query
-Structure paper
| Title | Transport and inhibition mechanism of the human SGLT2-MAP17 glucose transporter. |
|---|---|
| Journal, issue, pages | Nat Struct Mol Biol, Vol. 31, Issue 1, Page 159-169, Year 2024 |
| Publish date | Dec 6, 2023 |
 Authors Authors | Masahiro Hiraizumi / Tomoya Akashi / Kouta Murasaki / Hiroyuki Kishida / Taichi Kumanomidou / Nao Torimoto / Osamu Nureki / Ikuko Miyaguchi /  |
| PubMed Abstract | Sodium-glucose cotransporter 2 (SGLT2) is imporant in glucose reabsorption. SGLT2 inhibitors suppress renal glucose reabsorption, therefore reducing blood glucose levels in patients with type 2 ...Sodium-glucose cotransporter 2 (SGLT2) is imporant in glucose reabsorption. SGLT2 inhibitors suppress renal glucose reabsorption, therefore reducing blood glucose levels in patients with type 2 diabetes. We and others have developed several SGLT2 inhibitors starting from phlorizin, a natural product. Using cryo-electron microscopy, we present the structures of human (h)SGLT2-MAP17 complexed with five natural or synthetic inhibitors. The four synthetic inhibitors (including canagliflozin) bind the transporter in the outward conformations, while phlorizin binds it in the inward conformation. The phlorizin-hSGLT2 interaction exhibits biphasic kinetics, suggesting that phlorizin alternately binds to the extracellular and intracellular sides. The Na-bound outward-facing and unbound inward-open structures of hSGLT2-MAP17 suggest that the MAP17-associated bundle domain functions as a scaffold, with the hash domain rotating around the Na-binding site. Thus, Na binding stabilizes the outward-facing conformation, and its release promotes state transition to inward-open conformation, exhibiting a role of Na in symport mechanism. These results provide structural evidence for the Na-coupled alternating-access mechanism proposed for the transporter family. |
 External links External links |  Nat Struct Mol Biol / Nat Struct Mol Biol /  PubMed:38057552 / PubMed:38057552 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.8 - 3.3 Å |
| Structure data | EMDB-34610, PDB-8hb0: EMDB-34673, PDB-8hdh: EMDB-34705, PDB-8hez: EMDB-34737, PDB-8hg7: EMDB-34823, PDB-8hin: |
| Chemicals |  ChemComp-NAG: 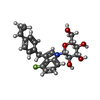 ChemComp-KZ3:  ChemComp-NA:  ChemComp-HOH:  ChemComp-L3R: 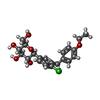 ChemComp-LE6: 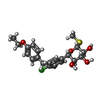 ChemComp-LFL:  ChemComp-LN9: |
| Source |
|
 Keywords Keywords |  TRANSPORT PROTEIN / TRANSPORT PROTEIN /  Ion transport / Sodium transport / Sugar transport / Ion transport / Sodium transport / Sugar transport /  Symport / Transport Symport / Transport |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers














