+Search query
-Structure paper
| Title | Structure and proposed DNA delivery mechanism of a marine roseophage. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 14, Issue 1, Page 3609, Year 2023 |
| Publish date | Jun 17, 2023 |
 Authors Authors | Yang Huang / Hui Sun / Shuzhen Wei / Lanlan Cai / Liqin Liu / Yanan Jiang / Jiabao Xin / Zhenqin Chen / Yuqiong Que / Zhibo Kong / Tingting Li / Hai Yu / Jun Zhang / Ying Gu / Qingbing Zheng / Shaowei Li / Rui Zhang / Ningshao Xia /  |
| PubMed Abstract | Tailed bacteriophages (order, Caudovirales) account for the majority of all phages. However, the long flexible tail of siphophages hinders comprehensive investigation of the mechanism of viral gene ...Tailed bacteriophages (order, Caudovirales) account for the majority of all phages. However, the long flexible tail of siphophages hinders comprehensive investigation of the mechanism of viral gene delivery. Here, we report the atomic capsid and in-situ structures of the tail machine of the marine siphophage, vB_DshS-R4C (R4C), which infects Roseobacter. The R4C virion, comprising 12 distinct structural protein components, has a unique five-fold vertex of the icosahedral capsid that allows genome delivery. The specific position and interaction pattern of the tail tube proteins determine the atypical long rigid tail of R4C, and further provide negative charge distribution within the tail tube. A ratchet mechanism assists in DNA transmission, which is initiated by an absorption device that structurally resembles the phage-like particle, RcGTA. Overall, these results provide in-depth knowledge into the intact structure and underlining DNA delivery mechanism for the ecologically important siphophages. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:37330604 / PubMed:37330604 /  PubMed Central PubMed Central |
| Methods | EM (single particle) / EM (helical sym.) |
| Resolution | 3.43 - 11.5 Å |
| Structure data | EMDB-34247, PDB-8gta: EMDB-34248, PDB-8gtb: EMDB-34249, PDB-8gtc: EMDB-34250, PDB-8gtd: EMDB-34252, PDB-8gtf: 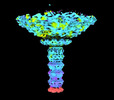 EMDB-34253: Dinoroseobacter phage vB_DshS-R4C C1 portal vertex  EMDB-34254: Dinoroseobacter phage vB_DshS-R4C C5 portal vertex |
| Source |
|
 Keywords Keywords |  VIRAL PROTEIN / Marine bacteriophage / Cyro-EM / Siphophage / VIRAL PROTEIN / Marine bacteriophage / Cyro-EM / Siphophage /  capsid / capsid /  Cryo-EM / tail tube protein / Cryo-EM / tail tube protein /  Baseplate / Megatron protein / Tail fibre protein / Distal tail protein / Hub protein / Baseplate / Megatron protein / Tail fibre protein / Distal tail protein / Hub protein /  VIRUS / Portal protein / Adaptor protein / Head-to-tail interface / Stopper protein / Terminator protein VIRUS / Portal protein / Adaptor protein / Head-to-tail interface / Stopper protein / Terminator protein |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers













 dinoroseobacter phage vb_dshs-r4c (virus)
dinoroseobacter phage vb_dshs-r4c (virus)