+Search query
-Structure paper
| Title | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding. |
|---|---|
| Journal, issue, pages | Cell, Vol. 185, Issue 7, Page 1143-1156.e13, Year 2022 |
| Publish date | Mar 31, 2022 |
 Authors Authors | Matthew Thomas Doyle / John R Jimah / Tyrone Dowdy / Shannon I Ohlemacher / Mioara Larion / Jenny E Hinshaw / Harris D Bernstein /  |
| PubMed Abstract | Transmembrane β barrel proteins are folded into the outer membrane (OM) of Gram-negative bacteria by the β barrel assembly machinery (BAM) via a poorly understood process that occurs without known ...Transmembrane β barrel proteins are folded into the outer membrane (OM) of Gram-negative bacteria by the β barrel assembly machinery (BAM) via a poorly understood process that occurs without known external energy sources. Here, we used single-particle cryo-EM to visualize the folding dynamics of a model β barrel protein (EspP) by BAM. We found that BAM binds the highly conserved "β signal" motif of EspP to correctly orient β strands in the OM during folding. We also found that the folding of EspP proceeds via "hybrid-barrel" intermediates in which membrane integrated β sheets are attached to the essential BAM subunit, BamA. The structures show an unprecedented deflection of the membrane surrounding the EspP intermediates and suggest that β sheets progressively fold toward BamA to form a β barrel. Along with in vivo experiments that tracked β barrel folding while the OM tension was modified, our results support a model in which BAM harnesses OM elasticity to accelerate β barrel folding. |
 External links External links |  Cell / Cell /  PubMed:35294859 / PubMed:35294859 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.6 - 4.8 Å |
| Structure data | EMDB-26105, PDB-7tsz: EMDB-26106, PDB-7tt0: EMDB-26107, PDB-7tt1: EMDB-26108, PDB-7tt2: EMDB-26109, PDB-7tt3: EMDB-26110, PDB-7tt4: EMDB-26111, PDB-7tt5: EMDB-26112, PDB-7tt6: EMDB-26113, PDB-7tt7: EMDB-26114, PDB-7ttc: |
| Chemicals | 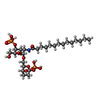 ChemComp-K33: |
| Source |
|
 Keywords Keywords |  MEMBRANE PROTEIN / MEMBRANE PROTEIN /  membrane protein folding / membrane dynamics / membrane protein folding / membrane dynamics /  outer membrane protein / BAM / outer membrane protein / BAM /  beta-barrel beta-barrel |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers