+Search query
-Structure paper
| Title | Structure of ATP synthase under strain during catalysis. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 13, Issue 1, Page 2232, Year 2022 |
| Publish date | Apr 25, 2022 |
 Authors Authors | Hui Guo / John L Rubinstein /  |
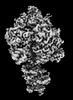
| PubMed Abstract | ATP synthases are macromolecular machines consisting of an ATP-hydrolysis-driven F motor and a proton-translocation-driven F motor. The F and F motors oppose each other's action on a shared rotor ...ATP synthases are macromolecular machines consisting of an ATP-hydrolysis-driven F motor and a proton-translocation-driven F motor. The F and F motors oppose each other's action on a shared rotor subcomplex and are held stationary relative to each other by a peripheral stalk. Structures of resting mitochondrial ATP synthases revealed a left-handed curvature of the peripheral stalk even though rotation of the rotor, driven by either ATP hydrolysis in F or proton translocation through F, would apply a right-handed bending force to the stalk. We used cryoEM to image yeast mitochondrial ATP synthase under strain during ATP-hydrolysis-driven rotary catalysis, revealing a large deformation of the peripheral stalk. The structures show how the peripheral stalk opposes the bending force and suggests that during ATP synthesis proton translocation causes accumulation of strain in the stalk, which relaxes by driving the relative rotation of the rotor through six sub-steps within F, leading to catalysis. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:35468906 / PubMed:35468906 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.2 - 8.4 Å |
| Structure data | EMDB-25930, PDB-7tjs: EMDB-25931, PDB-7tjt: EMDB-25932, PDB-7tju: EMDB-25933, PDB-7tjv: EMDB-25934, PDB-7tjw:  EMDB-25935: Yeast ATP synthase F1 region State 1catalytic beta_tight closed without exogenous ATP  EMDB-25936: Yeast ATP synthase F1 region State 1catalytic beta_tight open without exogenous ATP  EMDB-25937: Yeast ATP synthase F1 region State 1binding beta_tight open without exogenous ATP 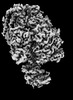 EMDB-25938: Yeast ATP synthase F1 region State 2catalytic beta_tight closed without exogenous ATP EMDB-25939, PDB-7tjx: 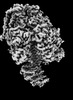 EMDB-25940: Yeast ATP synthase F1 region State 2catalytic beta_tight open without exogenous ATP  EMDB-25941: Yeast ATP synthase F1 region State 2binding beta_tight open without exogenous ATP  EMDB-25942: Yeast ATP synthase F1 region State 3catalytic beta_tight closed without exogenous ATP 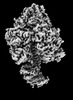 EMDB-25943: Yeast ATP synthase F1 region State 3catalytic beta_tight open without exogenous ATP  EMDB-25944: Yeast ATP synthase F1 region State 3binding beta_tight open without exogenous ATP  EMDB-25945: Yeast ATP synthase OSCP without exogenous ATP EMDB-25946, PDB-7tjy: EMDB-25947, PDB-7tjz: EMDB-25948, PDB-7tk0: EMDB-25949, PDB-7tk1:  EMDB-25950: Yeast ATP synthase peripheral stalk without exogenous ATP  EMDB-25951: Yeast ATP synthase FO region without exogenous ATP  EMDB-25952: Yeast ATP synthase composite map without exogenous ATP  EMDB-25953: Yeast ATP synthase with 10 mM ATP showing strained peripheral stalk determined by a screening microscope EMDB-25954, PDB-7tk2: EMDB-25955, PDB-7tk3: EMDB-25956, PDB-7tk4: EMDB-25957, PDB-7tk5: EMDB-25958, PDB-7tk6: EMDB-25959, PDB-7tk7: EMDB-25960, PDB-7tk8: EMDB-25961, PDB-7tk9: EMDB-25962, PDB-7tka: EMDB-25963, PDB-7tkb: EMDB-25964, PDB-7tkc: EMDB-25965, PDB-7tkd: EMDB-25966, PDB-7tke: EMDB-25967, PDB-7tkf: EMDB-25968, PDB-7tkg: EMDB-25969, PDB-7tkh: EMDB-25970, PDB-7tki: EMDB-25971, PDB-7tkj: EMDB-25972, PDB-7tkk: EMDB-25973, PDB-7tkl: EMDB-25974, PDB-7tkm: EMDB-25975, PDB-7tkn: EMDB-25976, PDB-7tko: EMDB-25977, PDB-7tkp: EMDB-25978, PDB-7tkq: EMDB-25979, PDB-7tkr: EMDB-25980, PDB-7tks: |
| Chemicals |  ChemComp-ATP:  ChemComp-MG:  ChemComp-ADP:  ChemComp-PO4: |
| Source |
|
 Keywords Keywords |  HYDROLASE / F1-ATPase / HYDROLASE / F1-ATPase /  ATP Synthase / ATP Synthase /  Nanomotor / Nanomotor /  Complex Complex |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers