+Search query
-Structure paper
| Title | Inactivation-mimicking block of the epithelial calcium channel TRPV6. |
|---|---|
| Journal, issue, pages | Sci Adv, Vol. 6, Issue 48, Year 2020 |
| Publish date | Nov 27, 2020 |
 Authors Authors | Rajesh Bhardwaj / Sonja Lindinger / Arthur Neuberger / Kirill D Nadezhdin / Appu K Singh / Micael R Cunha / Isabella Derler / Gergely Gyimesi / Jean-Louis Reymond / Matthias A Hediger / Christoph Romanin / Alexander I Sobolevsky /     |
| PubMed Abstract | Epithelial calcium channel TRPV6 plays vital roles in calcium homeostasis, and its dysregulation is implicated in multifactorial diseases, including cancers. Here, we study the molecular mechanism of ...Epithelial calcium channel TRPV6 plays vital roles in calcium homeostasis, and its dysregulation is implicated in multifactorial diseases, including cancers. Here, we study the molecular mechanism of selective nanomolar-affinity TRPV6 inhibition by (4-phenylcyclohexyl)piperazine derivatives (PCHPDs). We use x-ray crystallography and cryo-electron microscopy to solve the inhibitor-bound structures of TRPV6 and identify two types of inhibitor binding sites in the transmembrane region: (i) modulatory sites between the S1-S4 and pore domains normally occupied by lipids and (ii) the main site in the ion channel pore. Our structural data combined with mutagenesis, functional and computational approaches suggest that PCHPDs plug the open pore of TRPV6 and convert the channel into a nonconducting state, mimicking the action of calmodulin, which causes inactivation of TRPV6 channels under physiological conditions. This mechanism of inhibition explains the high selectivity and potency of PCHPDs and opens up unexplored avenues for the design of future-generation biomimetic drugs. |
 External links External links |  Sci Adv / Sci Adv /  PubMed:33246965 / PubMed:33246965 /  PubMed Central PubMed Central |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 3.1 - 4.34 Å |
| Structure data | EMDB-22662, PDB-7k4a: EMDB-22663, PDB-7k4b: EMDB-22664, PDB-7k4c: EMDB-22665, PDB-7k4d: EMDB-22666, PDB-7k4e: EMDB-22667, PDB-7k4f:  PDB-7d2k: |
| Chemicals |  ChemComp-CA: 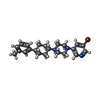 ChemComp-GQC:  ChemComp-Y01:  ChemComp-PCW: 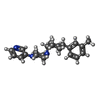 ChemComp-VUG: 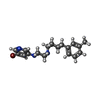 ChemComp-81F: 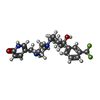 ChemComp-VUM:  ChemComp-VUJ:  ChemComp-5GK: |
| Source |
|
 Keywords Keywords | TRANSPORT PROTEIN/INHIBITOR /  Ion channels / Ion channels /  TRP channels / TRP channels /  Membrane proteins / TRANSPORT PROTEIN-INHIBITOR complex / Membrane proteins / TRANSPORT PROTEIN-INHIBITOR complex /  MEMBRANE PROTEIN / MEMBRANE PROTEIN /  TRPV6 / TRPV6 /  ion channel / open state / ion channel / open state /  MEMBRANE PROTEIN/INHIBITOR / MEMBRANE PROTEIN/INHIBITOR /  inhibitor / inhibitor /  MEMBRANE PROTEIN-INHIBITOR complex MEMBRANE PROTEIN-INHIBITOR complex |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers

















