+Search query
-Structure paper
| Title | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility. |
|---|---|
| Journal, issue, pages | Mol Cell, Vol. 82, Issue 7, Page 1278-11287.e5, Year 2022 |
| Publish date | Apr 7, 2022 |
 Authors Authors | Yan Chen / Yuwei Zhu / Xiang Li / Wenbo Gao / Ziqi Zhen / De Dong / Buliao Huang / Zhuo Ma / Anqi Zhang / Xiaocui Song / Yan Ma / Changyou Guo / Fan Zhang / Zhiwei Huang /  |
| PubMed Abstract | Cholesterol molecules specifically bind to the resting αβTCR to inhibit cytoplasmic CD3ζ ITAM phosphorylation through sequestering the TCR-CD3 complex in an inactive conformation. The mechanisms ...Cholesterol molecules specifically bind to the resting αβTCR to inhibit cytoplasmic CD3ζ ITAM phosphorylation through sequestering the TCR-CD3 complex in an inactive conformation. The mechanisms of cholesterol-mediated inhibition of TCR-CD3 and its activation remain unclear. Here, we present cryoelectron microscopy structures of cholesterol- and cholesterol sulfate (CS)-inhibited TCR-CD3 complexes and an auto-active TCR-CD3 variant. The structures reveal that cholesterol molecules act like a latch to lock CD3ζ into an inactive conformation in the membrane. Mutations impairing binding of cholesterol molecules to the tunnel result in the movement of the proximal C terminus of the CD3ζ transmembrane helix, thereby activating the TCR-CD3 complex in human cells. Together, our data reveal the structural basis of TCR inhibition by cholesterol, illustrate how the cholesterol-binding tunnel is allosterically coupled to TCR triggering, and lay a foundation for the development of immunotherapies through directly targeting the TCR-CD3 complex. |
 External links External links |  Mol Cell / Mol Cell /  PubMed:35271814 PubMed:35271814 |
| Methods | EM (single particle) |
| Resolution | 3.0 - 3.2 Å |
| Structure data | EMDB-31618, PDB-7fjd: EMDB-31619, PDB-7fje: EMDB-31620, PDB-7fjf: |
| Chemicals |  ChemComp-CLR: 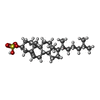 ChemComp-C3S: |
| Source |
|
 Keywords Keywords |  IMMUNE SYSTEM / IMMUNE SYSTEM /  membrane / membrane /  immune immune |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers










