+Search query
-Structure paper
| Title | Ligand and G-protein selectivity in the κ-opioid receptor. |
|---|---|
| Journal, issue, pages | Nature, Vol. 617, Issue 7960, Page 417-425, Year 2023 |
| Publish date | May 3, 2023 |
 Authors Authors | Jianming Han / Jingying Zhang / Antonina L Nazarova / Sarah M Bernhard / Brian E Krumm / Lei Zhao / Jordy Homing Lam / Vipin A Rangari / Susruta Majumdar / David E Nichols / Vsevolod Katritch / Peng Yuan / Jonathan F Fay / Tao Che /  |
| PubMed Abstract | The κ-opioid receptor (KOR) represents a highly desirable therapeutic target for treating not only pain but also addiction and affective disorders. However, the development of KOR analgesics has ...The κ-opioid receptor (KOR) represents a highly desirable therapeutic target for treating not only pain but also addiction and affective disorders. However, the development of KOR analgesics has been hindered by the associated hallucinogenic side effects. The initiation of KOR signalling requires the G-family proteins including the conventional (G, G, G, G and G) and nonconventional (G and G) subtypes. How hallucinogens exert their actions through KOR and how KOR determines G-protein subtype selectivity are not well understood. Here we determined the active-state structures of KOR in a complex with multiple G-protein heterotrimers-G, G, G and G-using cryo-electron microscopy. The KOR-G-protein complexes are bound to hallucinogenic salvinorins or highly selective KOR agonists. Comparisons of these structures reveal molecular determinants critical for KOR-G-protein interactions as well as key elements governing G-family subtype selectivity and KOR ligand selectivity. Furthermore, the four G-protein subtypes display an intrinsically different binding affinity and allosteric activity on agonist binding at KOR. These results provide insights into the actions of opioids and G-protein-coupling specificity at KOR and establish a foundation to examine the therapeutic potential of pathway-selective agonists of KOR. |
 External links External links |  Nature / Nature /  PubMed:37138078 / PubMed:37138078 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.61 - 2.82 Å |
| Structure data | EMDB-27804, PDB-8dzp: EMDB-27805, PDB-8dzq: EMDB-27806, PDB-8dzr: EMDB-27807, PDB-8dzs:  EMDB-41876: momSalB bound Kappa Opioid Receptor in complex with GoA 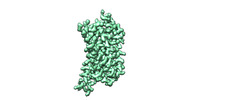 EMDB-43732: momSalB bound Kappa Opioid Receptor in complex Gi1  EMDB-43733: GR89,696 bound Kappa Opioid Receptor in complex with Gz  EMDB-43734: GR89,696 bound Kappa Opioid Receptor in complex with gustducin |
| Chemicals | 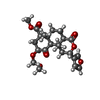 ChemComp-U99: 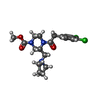 ChemComp-U9I: |
| Source |
|
 Keywords Keywords |  MEMBRANE PROTEIN / MEMBRANE PROTEIN /  GPCR / GPCR /  KAPPA OPIOID Receptor / SALVINORIN / Receptor / KAPPA OPIOID Receptor / SALVINORIN / Receptor /  Kappa / KOR / Kappa / KOR /  Opioid / Opioid /  G protein G protein |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers













