+Search query
-Structure paper
| Title | Molecular basis of the pleiotropic effects by the antibiotic amikacin on the ribosome. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 14, Issue 1, Page 4666, Year 2023 |
| Publish date | Aug 3, 2023 |
 Authors Authors | Savannah M Seely / Narayan P Parajuli / Arindam De Tarafder / Xueliang Ge / Suparna Sanyal / Matthieu G Gagnon /   |
| PubMed Abstract | Aminoglycosides are a class of antibiotics that bind to ribosomal RNA and exert pleiotropic effects on ribosome function. Amikacin, the semisynthetic derivative of kanamycin, is commonly used for ...Aminoglycosides are a class of antibiotics that bind to ribosomal RNA and exert pleiotropic effects on ribosome function. Amikacin, the semisynthetic derivative of kanamycin, is commonly used for treating severe infections with multidrug-resistant, aerobic Gram-negative bacteria. Amikacin carries the 4-amino-2-hydroxy butyrate (AHB) moiety at the N amino group of the central 2-deoxystreptamine (2-DOS) ring, which may confer amikacin a unique ribosome inhibition profile. Here we use in vitro fast kinetics combined with X-ray crystallography and cryo-EM to dissect the mechanisms of ribosome inhibition by amikacin and the parent compound, kanamycin. Amikacin interferes with tRNA translocation, release factor-mediated peptidyl-tRNA hydrolysis, and ribosome recycling, traits attributed to the additional interactions amikacin makes with the decoding center. The binding site in the large ribosomal subunit proximal to the 3'-end of tRNA in the peptidyl (P) site lays the groundwork for rational design of amikacin derivatives with improved antibacterial properties. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:37537169 / PubMed:37537169 /  PubMed Central PubMed Central |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 2.89 - 2.946 Å |
| Structure data | EMDB-40882, PDB-8syl:  PDB-8ev6:  PDB-8ev7: |
| Chemicals |  ChemComp-MG: 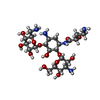 ChemComp-AKN:  ChemComp-ZN:  ChemComp-SF4:  ChemComp-HOH:  ChemComp-KAN:  ChemComp-PHE: |
| Source |
|
 Keywords Keywords |  RIBOSOME / RIBOSOME /  Amikacin / Amikacin /  antibiotic / antibiotic /  aminoglycoside / aminoglycoside /  70S ribosome / 70S ribosome /  inhibitor / inhibitor /  tRNA / tRNA /  Kanamycin / Kanamycin /  cryo-EM cryo-EM |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers