+Search query
-Structure paper
| Title | Ligand activation mechanisms of human KCNQ2 channel. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 14, Issue 1, Page 6632, Year 2023 |
| Publish date | Oct 19, 2023 |
 Authors Authors | Demin Ma / Yueming Zheng / Xiaoxiao Li / Xiaoyu Zhou / Zhenni Yang / Yan Zhang / Long Wang / Wenbo Zhang / Jiajia Fang / Guohua Zhao / Panpan Hou / Fajun Nan / Wei Yang / Nannan Su / Zhaobing Gao / Jiangtao Guo /  |
| PubMed Abstract | The human voltage-gated potassium channel KCNQ2/KCNQ3 carries the neuronal M-current, which helps to stabilize the membrane potential. KCNQ2 can be activated by analgesics and antiepileptic drugs but ...The human voltage-gated potassium channel KCNQ2/KCNQ3 carries the neuronal M-current, which helps to stabilize the membrane potential. KCNQ2 can be activated by analgesics and antiepileptic drugs but their activation mechanisms remain unclear. Here we report cryo-electron microscopy (cryo-EM) structures of human KCNQ2-CaM in complex with three activators, namely the antiepileptic drug cannabidiol (CBD), the lipid phosphatidylinositol 4,5-bisphosphate (PIP), and HN37 (pynegabine), an antiepileptic drug in the clinical trial, in an either closed or open conformation. The activator-bound structures, along with electrophysiology analyses, reveal the binding modes of two CBD, one PIP, and two HN37 molecules in each KCNQ2 subunit, and elucidate their activation mechanisms on the KCNQ2 channel. These structures may guide the development of antiepileptic drugs and analgesics that target KCNQ2. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:37857637 / PubMed:37857637 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.5 - 3.5 Å |
| Structure data | EMDB-35877, PDB-8izy: EMDB-35879, PDB-8j00: EMDB-35880, PDB-8j01: EMDB-35881, PDB-8j02: EMDB-35882, PDB-8j03: EMDB-35883, PDB-8j04: EMDB-35884, PDB-8j05: EMDB-37270, PDB-8w4u: |
| Chemicals | 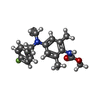 ChemComp-9MF: 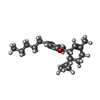 ChemComp-P0T:  ChemComp-PIO: |
| Source |
|
 Keywords Keywords |  MEMBRANE PROTEIN / MEMBRANE PROTEIN /  potassium voltage-gated channel / HN37 / CBD / potassium voltage-gated channel / HN37 / CBD /  PIP2 PIP2 |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




















