+Search query
-Structure paper
| Title | Structural and functional investigations of syn-copalyl diphosphate synthase from Oryza sativa. |
|---|---|
| Journal, issue, pages | Commun Chem, Vol. 6, Issue 1, Page 240, Year 2023 |
| Publish date | Nov 6, 2023 |
 Authors Authors | Xiaoli Ma / Haifeng Xu / Yuru Tong / Yunfeng Luo / Qinghua Dong / Tao Jiang /  |
| PubMed Abstract | The large superfamily of labdane-related diterpenoids is defined by the cyclization of linear geranylgeranyl pyrophosphate (GGPP), catalyzed by copalyl diphosphate synthases (CPSs) to form the basic ...The large superfamily of labdane-related diterpenoids is defined by the cyclization of linear geranylgeranyl pyrophosphate (GGPP), catalyzed by copalyl diphosphate synthases (CPSs) to form the basic decalin core, the copalyl diphosphates (CPPs). Three stereochemically distinct CPPs have been found in plants, namely (+)-CPP, ent-CPP and syn-CPP. Here, we used X-ray crystallography and cryo-EM methods to describe different oligomeric structures of a syn-copalyl diphosphate synthase from Oryza sativa (OsCyc1), and provided a cryo-EM structure of OsCyc1 mutant in complex with the substrate GGPP. Further analysis showed that tetramers are the dominant form of OsCyc1 in solution and are not necessary for enzyme activity in vitro. Through rational design, we identified an OsCyc1 mutant that can generate ent-CPP in addition to syn-CPP. Our work provides a structural and mechanistic basis for comparing different CPSs and paves the way for further enzyme design to obtain diterpene derivatives with specific chirality. |
 External links External links |  Commun Chem / Commun Chem /  PubMed:37932442 / PubMed:37932442 /  PubMed Central PubMed Central |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 3.49 - 7.9 Å |
| Structure data | EMDB-35202, PDB-8i6p: EMDB-35206, PDB-8i6t: EMDB-35207, PDB-8i6u: EMDB-35440, PDB-8ih5:  PDB-8kbw: |
| Chemicals | 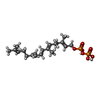 ChemComp-GRG: |
| Source |
|
 Keywords Keywords |  ISOMERASE / ISOMERASE /  syn-copalyl diphosphate synthase / labdane-related diterpenoids / oligomer cryo-EM structure / plant defense / syn-copalyl diphosphate synthase / labdane-related diterpenoids / oligomer cryo-EM structure / plant defense /  PLANT PROTEIN / hexamer cryo-EM structure / dimer cryo-EM structure / labdane-related diterpenoid / GGPP complexed cryo-EM structure / apo-state crystal structure PLANT PROTEIN / hexamer cryo-EM structure / dimer cryo-EM structure / labdane-related diterpenoid / GGPP complexed cryo-EM structure / apo-state crystal structure |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers












