+Search query
-Structure paper
| Title | Molecular basis of receptor binding and antibody neutralization of Omicron. |
|---|---|
| Journal, issue, pages | Nature, Vol. 604, Issue 7906, Page 546-552, Year 2022 |
| Publish date | Feb 28, 2022 |
 Authors Authors | Qin Hong / Wenyu Han / Jiawei Li / Shiqi Xu / Yifan Wang / Cong Xu / Zuyang Li / Yanxing Wang / Chao Zhang / Zhong Huang / Yao Cong /  |
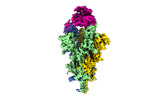
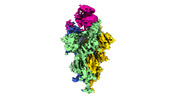
| PubMed Abstract | The SARS-CoV-2 Omicron variant exhibits striking immune evasion and is spreading rapidly worldwide. Understanding the structural basis of the high transmissibility and enhanced immune evasion of ...The SARS-CoV-2 Omicron variant exhibits striking immune evasion and is spreading rapidly worldwide. Understanding the structural basis of the high transmissibility and enhanced immune evasion of Omicron is of high importance. Here, using cryo-electron microscopy, we present both the closed and the open states of the Omicron spike (S) protein, which appear more compact than the counterparts of the G614 strain, potentially related to enhanced inter-protomer and S1-S2 interactions induced by Omicron residue substitution. The closed state showing dominant population may indicate a conformational masking mechanism for the immune evasion of Omicron. Moreover, we captured three states for the Omicron S-ACE2 complex, revealing that the substitutions on the Omicron RBM result in new salt bridges and hydrogen bonds, more favourable electrostatic surface properties, and an overall strengthened S-ACE2 interaction, in line with the observed higher ACE2 affinity of Omicron S than of G614. Furthermore, we determined the structures of Omicron S in complex with the Fab of S3H3, an antibody that is able to cross-neutralize major variants of concern including Omicron, elucidating the structural basis for S3H3-mediated broad-spectrum neutralization. Our findings shed light on the receptor engagement and antibody neutralization or evasion of Omicron and may also inform the design of broadly effective vaccines against SARS-CoV-2. |
 External links External links |  Nature / Nature /  PubMed:35228716 PubMed:35228716 |
| Methods | EM (single particle) |
| Resolution | 3.1 - 4.04 Å |
| Structure data | EMDB-32556, PDB-7wk2: EMDB-32557, PDB-7wk3: EMDB-32558, PDB-7wk4: EMDB-32559, PDB-7wk5: EMDB-32560, PDB-7wk6: EMDB-32562, PDB-7wk8: EMDB-32563, PDB-7wk9: EMDB-32564, PDB-7wka: EMDB-32854, PDB-7wvn: EMDB-32855, PDB-7wvo: EMDB-32856, PDB-7wvp: EMDB-32857, PDB-7wvq: |
| Source |
|
 Keywords Keywords |  VIRAL PROTEIN / VIRAL PROTEIN /  SARS-CoV-2 / SARS-CoV-2 /  coronavirus / coronavirus /  Omicron variant / S-close / Omicron variant / S-close /  spike protein / S-open / HYDROLASE/VIRAL PROTEIN / spike protein / S-open / HYDROLASE/VIRAL PROTEIN /  Omicron / Omicron /  ACE2 / HYDROLASE-VIRAL PROTEIN complex / ACE2 / HYDROLASE-VIRAL PROTEIN complex /  VIRAL PROTEIN/HYDROLASE / VIRAL PROTEIN/HYDROLASE /  VIRAL PROTEIN-HYDROLASE complex / VIRAL PROTEIN-HYDROLASE complex /  B.1.1.529 lineage / S-open-2 B.1.1.529 lineage / S-open-2 |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers