+Search query
-Structure paper
| Title | Agonists and allosteric modulators promote signaling from different metabotropic glutamate receptor 5 conformations. |
|---|---|
| Journal, issue, pages | Cell Rep, Vol. 36, Issue 9, Page 109648, Year 2021 |
| Publish date | Aug 31, 2021 |
 Authors Authors | Chady Nasrallah / Giuseppe Cannone / Julie Briot / Karine Rottier / Alice E Berizzi / Chia-Ying Huang / Robert B Quast / Francois Hoh / Jean-Louis Banères / Fanny Malhaire / Ludovic Berto / Anaëlle Dumazer / Joan Font-Ingles / Xavier Gómez-Santacana / Juanlo Catena / Julie Kniazeff / Cyril Goudet / Amadeu Llebaria / Jean-Philippe Pin / Kutti R Vinothkumar / Guillaume Lebon /      |
| PubMed Abstract | Metabotropic glutamate receptors (mGluRs) are dimeric G-protein-coupled receptors activated by the main excitatory neurotransmitter, L-glutamate. mGluR activation by agonists binding in the venus ...Metabotropic glutamate receptors (mGluRs) are dimeric G-protein-coupled receptors activated by the main excitatory neurotransmitter, L-glutamate. mGluR activation by agonists binding in the venus flytrap domain is regulated by positive (PAM) or negative (NAM) allosteric modulators binding to the 7-transmembrane domain (7TM). We report the cryo-electron microscopy structures of fully inactive and intermediate-active conformations of mGlu receptor bound to an antagonist and a NAM or an agonist and a PAM, respectively, as well as the crystal structure of the 7TM bound to a photoswitchable NAM. The agonist induces a large movement between the subunits, bringing the 7TMs together and stabilizing a 7TM conformation structurally similar to the inactive state. Using functional approaches, we demonstrate that the PAM stabilizes a 7TM active conformation independent of the conformational changes induced by agonists, representing an alternative mode of mGlu activation. These findings provide a structural basis for different mGluR activation modes. |
 External links External links |  Cell Rep / Cell Rep /  PubMed:34469715 / PubMed:34469715 /  PubMed Central PubMed Central |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 2.54 - 4.0 Å |
| Structure data | EMDB-31536, PDB-7fd8: EMDB-31537, PDB-7fd9:  PDB-7p2l: |
| Chemicals |  ChemComp-NAG: 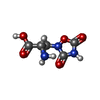 ChemComp-QUS:  ChemComp-Y01: 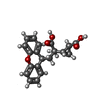 ChemComp-Z99: 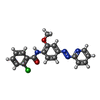 ChemComp-4YI:  ChemComp-HOH: |
| Source |
|
 Keywords Keywords |  MEMBRANE PROTEIN / G protein coupled receptors / MEMBRANE PROTEIN / G protein coupled receptors /  Signal transduction / Metabotropic glutamate receptor 5 (GRM5) / G-protein coupled receptors / Signal transduction / Metabotropic glutamate receptor 5 (GRM5) / G-protein coupled receptors /  metabotropic glutamate receptor / inactive state / metabotropic glutamate receptor / inactive state /  SIGNALING PROTEIN / SIGNALING PROTEIN /  G protein coupled receptor / mGlu5 7TM / photoswitchable ligand / G protein coupled receptor / mGlu5 7TM / photoswitchable ligand /  allosteric modulator allosteric modulator |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers









