+Search query
-Structure paper
| Title | Bivalent intra-spike binding provides durability against emergent Omicron lineages: Results from a global consortium. |
|---|---|
| Journal, issue, pages | Cell Rep, Vol. 42, Issue 1, Page 112014, Year 2023 |
| Publish date | Jan 31, 2023 |
 Authors Authors | Heather M Callaway / Kathryn M Hastie / Sharon L Schendel / Haoyang Li / Xiaoying Yu / Jeremy Shek / Tierra Buck / Sean Hui / Dan Bedinger / Camille Troup / S Moses Dennison / Kan Li / Michael D Alpert / Charles C Bailey / Sharon Benzeno / Jody L Bonnevier / Jin-Qiu Chen / Charm Chen / Hyeseon Cho / Peter D Crompton / Vincent Dussupt / Kevin C Entzminger / Yassine Ezzyat / Jonathan K Fleming / Nick Geukens / Amy E Gilbert / Yongjun Guan / Xiaojian Han / Christopher J Harvey / Julia M Hatler / Bryan Howie / Chao Hu / Ailong Huang / Maya Imbrechts / Aishun Jin / Nik Kamachi / Gladys Keitany / Mark Klinger / Jay K Kolls / Shelly J Krebs / Tingting Li / Feiyan Luo / Toshiaki Maruyama / Michael A Meehl / Letzibeth Mendez-Rivera / Andrea Musa / C J Okumura / Benjamin E R Rubin / Aaron K Sato / Meiying Shen / Anirudh Singh / Shuyi Song / Joshua Tan / Jeffrey M Trimarchi / Dhruvkumar P Upadhyay / Yingming Wang / Lei Yu / Tom Z Yuan / Erik Yusko / Bjoern Peters / Georgia Tomaras / Erica Ollmann Saphire /     |
| PubMed Abstract | The SARS-CoV-2 Omicron variant of concern (VoC) and its sublineages contain 31-36 mutations in spike and escape neutralization by most therapeutic antibodies. In a pseudovirus neutralization assay, ...The SARS-CoV-2 Omicron variant of concern (VoC) and its sublineages contain 31-36 mutations in spike and escape neutralization by most therapeutic antibodies. In a pseudovirus neutralization assay, 66 of the nearly 400 candidate therapeutics in the Coronavirus Immunotherapeutic Consortium (CoVIC) panel neutralize Omicron and multiple Omicron sublineages. Among natural immunoglobulin Gs (IgGs), especially those in the receptor-binding domain (RBD)-2 epitope community, nearly all Omicron neutralizers recognize spike bivalently, with both antigen-binding fragments (Fabs) simultaneously engaging adjacent RBDs on the same spike. Most IgGs that do not neutralize Omicron bind either entirely monovalently or have some (22%-50%) monovalent occupancy. Cleavage of bivalent-binding IgGs to Fabs abolishes neutralization and binding affinity, with disproportionate loss of activity against Omicron pseudovirus and spike. These results suggest that VoC-resistant antibodies overcome mutagenic substitution via avidity. Hence, vaccine strategies targeting future SARS-CoV-2 variants should consider epitope display with spacing and organization identical to trimeric spike. |
 External links External links |  Cell Rep / Cell Rep /  PubMed:36681898 / PubMed:36681898 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 16.0 - 26.0 Å |
| Structure data |  EMDB-28090: Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-040  EMDB-28091: Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-045  EMDB-28092: Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-093 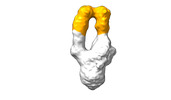 EMDB-28093: Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-156 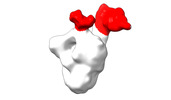 EMDB-28094: Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-234  EMDB-28095: Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-260  EMDB-28096: Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-279  EMDB-28097: Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-290 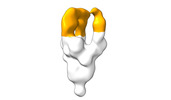 EMDB-28098: Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-294  EMDB-28099: Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-295  EMDB-28100: Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-299  EMDB-28102: Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-334 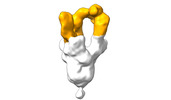 EMDB-28103: Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-360  EMDB-28104: Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-361  EMDB-28105: Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-362  EMDB-28106: Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-368  EMDB-28168: Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-292  EMDB-28169: Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-333  EMDB-28170: Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-355  EMDB-28171: Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-371 |
| Source |
|
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




 Severe acute respiratory syndrome coronavirus 2
Severe acute respiratory syndrome coronavirus 2