+Search query
-Structure paper
| Title | Structural consequences of turnover-induced homocitrate loss in nitrogenase. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 14, Issue 1, Page 1091, Year 2023 |
| Publish date | Feb 25, 2023 |
 Authors Authors | Rebeccah A Warmack / Ailiena O Maggiolo / Andres Orta / Belinda B Wenke / James B Howard / Douglas C Rees /  |
| PubMed Abstract | Nitrogenase catalyzes the ATP-dependent reduction of dinitrogen to ammonia during the process of biological nitrogen fixation that is essential for sustaining life. The active site FeMo-cofactor ...Nitrogenase catalyzes the ATP-dependent reduction of dinitrogen to ammonia during the process of biological nitrogen fixation that is essential for sustaining life. The active site FeMo-cofactor contains a [7Fe:1Mo:9S:1C] metallocluster coordinated with an R-homocitrate (HCA) molecule. Here, we establish through single particle cryoEM and chemical analysis of two forms of the Azotobacter vinelandii MoFe-protein - a high pH turnover inactivated species and a ∆NifV variant that cannot synthesize HCA - that loss of HCA is coupled to α-subunit domain and FeMo-cofactor disordering, and formation of a histidine coordination site. We further find a population of the ∆NifV variant complexed to an endogenous protein identified through structural and proteomic approaches as the uncharacterized protein NafT. Recognition by endogenous NafT demonstrates the physiological relevance of the HCA-compromised form, perhaps for cofactor insertion or repair. Our results point towards a dynamic active site in which HCA plays a role in enabling nitrogenase catalysis by facilitating activation of the FeMo-cofactor from a relatively stable form to a state capable of reducing dinitrogen under ambient conditions. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:36841829 / PubMed:36841829 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 1.92 - 2.71 Å |
| Structure data | EMDB-26957, PDB-8crs: EMDB-27316, PDB-8dbx: EMDB-28272, PDB-8enl: EMDB-28273, PDB-8enm: EMDB-28274, PDB-8enn: EMDB-28275, PDB-8eno: |
| Chemicals | 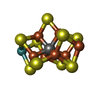 ChemComp-ICS: 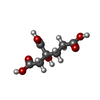 ChemComp-HCA: 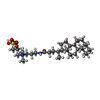 ChemComp-1N7:  ChemComp-CLF:  ChemComp-FE:  ChemComp-HOH:  ChemComp-1CL:  ChemComp-UNX:  ChemComp-CIT: |
| Source |
|
 Keywords Keywords | METAL BINDING PROTEIN /  OXIDOREDUCTASE / OXIDOREDUCTASE /  Nitrogenase / Nitrogenase /  metalloenzyme / metalloenzyme /  nitrogen fixation / nitrogen fixation /  reductase / MoFe reductase / MoFe |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers
















