+Search query
-Structure paper
| Title | Structural mechanism of glutamate receptor activation and desensitization. |
|---|---|
| Journal, issue, pages | Nature, Vol. 514, Issue 7522, Page 328-334, Year 2014 |
| Publish date | Oct 16, 2014 |
 Authors Authors | Joel R Meyerson / Janesh Kumar / Sagar Chittori / Prashant Rao / Jason Pierson / Alberto Bartesaghi / Mark L Mayer / Sriram Subramaniam /  |
| PubMed Abstract | Ionotropic glutamate receptors are ligand-gated ion channels that mediate excitatory synaptic transmission in the vertebrate brain. To gain a better understanding of how structural changes gate ion ...Ionotropic glutamate receptors are ligand-gated ion channels that mediate excitatory synaptic transmission in the vertebrate brain. To gain a better understanding of how structural changes gate ion flux across the membrane, we trapped rat AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid) and kainate receptor subtypes in their major functional states and analysed the resulting structures using cryo-electron microscopy. We show that transition to the active state involves a 'corkscrew' motion of the receptor assembly, driven by closure of the ligand-binding domain. Desensitization is accompanied by disruption of the amino-terminal domain tetramer in AMPA, but not kainate, receptors with a two-fold to four-fold symmetry transition in the ligand-binding domains in both subtypes. The 7.6 Å structure of a desensitized kainate receptor shows how these changes accommodate channel closing. These findings integrate previous physiological, biochemical and structural analyses of glutamate receptors and provide a molecular explanation for key steps in receptor gating. |
 External links External links |  Nature / Nature /  PubMed:25119039 / PubMed:25119039 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 7.6 - 25.9 Å |
| Structure data | EMDB-2680: Density map of GluA2em in complex with ZK200775 EMDB-2684: Density map of GluA2em in complex with LY451646 and glutamate EMDB-2685: Density map of GluK2 desensitized state in complex with 2S,4R-4-methylglutamate  EMDB-2686:  EMDB-2687:  EMDB-2688: |
| Chemicals |  ChemComp-GLU: 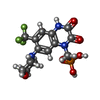 ChemComp-ZK1: 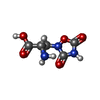 ChemComp-QUS: |
| Source |
|
 Keywords Keywords |  TRANSPORT PROTEIN / TRANSPORT PROTEIN /  MEMBRANE PROTEIN / MEMBRANE PROTEIN /  ION CHANNEL / GLUA2EM ANTAGONIST-BOUND CLOSED STATE / GLUA2EM RESTORED ACTIVE STATE ION CHANNEL / GLUA2EM ANTAGONIST-BOUND CLOSED STATE / GLUA2EM RESTORED ACTIVE STATE |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers












