+Search query
-Structure paper
| Title | New insights into rotavirus entry machinery: stabilization of rotavirus spike conformation is independent of trypsin cleavage. |
|---|---|
| Journal, issue, pages | PLoS Pathog, Vol. 10, Issue 5, Page e1004157, Year 2014 |
| Publish date | May 29, 2014 |
 Authors Authors | Javier M Rodríguez / Francisco J Chichón / Esther Martín-Forero / Fernando González-Camacho / José L Carrascosa / José R Castón / Daniel Luque /  |
| PubMed Abstract | The infectivity of rotavirus, the main causative agent of childhood diarrhea, is dependent on activation of the extracellular viral particles by trypsin-like proteases in the host intestinal lumen. ...The infectivity of rotavirus, the main causative agent of childhood diarrhea, is dependent on activation of the extracellular viral particles by trypsin-like proteases in the host intestinal lumen. This step entails proteolytic cleavage of the VP4 spike protein into its mature products, VP8* and VP5*. Previous cryo-electron microscopy (cryo-EM) analysis of trypsin-activated particles showed well-resolved spikes, although no density was identified for the spikes in uncleaved particles; these data suggested that trypsin activation triggers important conformational changes that give rise to the rigid, entry-competent spike. The nature of these structural changes is not well understood, due to lack of data relative to the uncleaved spike structure. Here we used cryo-EM and cryo-electron tomography (cryo-ET) to characterize the structure of the uncleaved virion in two model rotavirus strains. Cryo-EM three-dimensional reconstruction of uncleaved virions showed spikes with a structure compatible with the atomic model of the cleaved spike, and indistinguishable from that of digested particles. Cryo-ET and subvolume average, combined with classification methods, resolved the presence of non-icosahedral structures, providing a model for the complete structure of the uncleaved spike. Despite the similar rigid structure observed for uncleaved and cleaved particles, trypsin activation is necessary for successful infection. These observations suggest that the spike precursor protein must be proteolytically processed, not to achieve a rigid conformation, but to allow the conformational changes that drive virus entry. |
 External links External links |  PLoS Pathog / PLoS Pathog /  PubMed:24873828 / PubMed:24873828 /  PubMed Central PubMed Central |
| Methods | EM (single particle) / EM (subtomogram averaging) |
| Resolution | 14.5 - 17.4 Å |
| Structure data |  EMDB-2573:  EMDB-2574:  EMDB-2575:  EMDB-2576:  EMDB-2577: 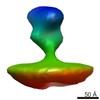 EMDB-2578:  EMDB-2579: 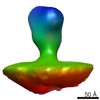 EMDB-2580: |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



