+Search query
-Structure paper
| Title | Structure and desensitization of AMPA receptor complexes with type II TARP γ5 and GSG1L. |
|---|---|
| Journal, issue, pages | Mol Cell, Vol. 81, Issue 23, Page 4771-44783.e7, Year 2021 |
| Publish date | Dec 2, 2021 |
 Authors Authors | Oleg Klykov / Shanti Pal Gangwar / Maria V Yelshanskaya / Laura Yen / Alexander I Sobolevsky /  |
| PubMed Abstract | AMPA receptors (AMPARs) mediate the majority of excitatory neurotransmission. Their surface expression, trafficking, gating, and pharmacology are regulated by auxiliary subunits. Of the two types of ...AMPA receptors (AMPARs) mediate the majority of excitatory neurotransmission. Their surface expression, trafficking, gating, and pharmacology are regulated by auxiliary subunits. Of the two types of TARP auxiliary subunits, type I TARPs assume activating roles, while type II TARPs serve suppressive functions. We present cryo-EM structures of GluA2 AMPAR in complex with type II TARP γ5, which reduces steady-state currents, increases single-channel conductance, and slows recovery from desensitization. Regulation of AMPAR function depends on its ligand-binding domain (LBD) interaction with the γ5 head domain. GluA2-γ5 complex shows maximum stoichiometry of two TARPs per AMPAR tetramer, being different from type I TARPs but reminiscent of the auxiliary subunit GSG1L. Desensitization of both GluA2-GSG1L and GluA2-γ5 complexes is accompanied by rupture of LBD dimer interface, while GluA2-γ5 but not GluA2-GSG1L LBD dimers remain two-fold symmetric. Different structural architectures and desensitization mechanisms of complexes with auxiliary subunits endow AMPARs with broad functional capabilities. |
 External links External links |  Mol Cell / Mol Cell /  PubMed:34678168 / PubMed:34678168 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.3 - 4.4 Å |
| Structure data | EMDB-24748, PDB-7ryy: EMDB-24749, PDB-7ryz: EMDB-24750, PDB-7rz4: EMDB-24751, PDB-7rz5: EMDB-24752, PDB-7rz6: EMDB-24753, PDB-7rz7: EMDB-24754, PDB-7rz8: EMDB-24755, PDB-7rz9: EMDB-24756, PDB-7rza: |
| Chemicals |  ChemComp-PCW:  ChemComp-GLU: 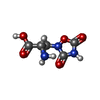 ChemComp-QUS: 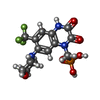 ChemComp-ZK1:  ChemComp-NAG: |
| Source |
|
 Keywords Keywords |  MEMBRANE PROTEIN / MEMBRANE PROTEIN /  AMPA receptor / AMPA receptor /  ion channel / ion channel /  neurotransmission / neurotransmission /  synapse / TARP gamma-5 / GSG1L synapse / TARP gamma-5 / GSG1L |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers






















