+Search query
-Structure paper
| Title | Local Stabilization of Subunit-Subunit Contacts Causes Global Destabilization of Hepatitis B Virus Capsids. |
|---|---|
| Journal, issue, pages | ACS Chem Biol, Vol. 15, Issue 6, Page 1708-1717, Year 2020 |
| Publish date | Jun 19, 2020 |
 Authors Authors | Christopher John Schlicksup / Patrick Laughlin / Steven Dunkelbarger / Joseph Che-Yen Wang / Adam Zlotnick /  |
| PubMed Abstract | Development of antiviral molecules that bind virion is a strategy that remains in its infancy, and the details of their mechanisms are poorly understood. Here we investigate the behavior of DBT1, a ...Development of antiviral molecules that bind virion is a strategy that remains in its infancy, and the details of their mechanisms are poorly understood. Here we investigate the behavior of DBT1, a dibenzothiazepine that specifically interacts with the capsid protein of hepatitis B virus (HBV). We found that DBT1 stabilizes protein-protein interaction, accelerates capsid assembly, and can induce formation of aberrant particles. Paradoxically, DBT1 can cause preformed capsids to dissociate. These activities may lead to (i) assembly of empty and defective capsids, inhibiting formation of new virus, and (ii) disruption of mature viruses, which are metastable, to inhibit new infection. Using cryo-electron microscopy, we observed that DBT1 led to asymmetric capsids where well-defined DBT1 density was bound at all intersubunit contacts. These results suggest that DBT1 can support assembly by increasing buried surface area but induce disassembly of metastable capsids by favoring asymmetry to induce structural defects. |
 External links External links |  ACS Chem Biol / ACS Chem Biol /  PubMed:32369333 / PubMed:32369333 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.7 - 4.6 Å |
| Structure data | EMDB-21653, PDB-6wfs:  EMDB-21654:  EMDB-21655: |
| Chemicals | 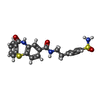 ChemComp-U0A: |
| Source |
|
 Keywords Keywords |  VIRUS LIKE PARTICLE / VIRUS LIKE PARTICLE /  capsid / capsid /  antiviral / HBV antiviral / HBV |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers






