+Search query
-Structure paper
| Title | ABCG2 transports anticancer drugs via a closed-to-open switch. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 11, Issue 1, Page 2264, Year 2020 |
| Publish date | May 8, 2020 |
 Authors Authors | Benjamin J Orlando / Maofu Liao /  |
| PubMed Abstract | ABCG2 is an ABC transporter that extrudes a variety of compounds from cells, and presents an obstacle in treating chemotherapy-resistant cancers. Despite recent structural insights, no anticancer ...ABCG2 is an ABC transporter that extrudes a variety of compounds from cells, and presents an obstacle in treating chemotherapy-resistant cancers. Despite recent structural insights, no anticancer drug bound to ABCG2 has been resolved, and the mechanisms of multidrug transport remain obscure. Such a gap of knowledge limits the development of novel compounds that block or evade this critical molecular pump. Here we present single-particle cryo-EM studies of ABCG2 in the apo state, and bound to the three structurally distinct chemotherapeutics. Without the binding of conformation-selective antibody fragments or inhibitors, the resting ABCG2 adopts a closed conformation. Our cryo-EM, biochemical, and functional analyses reveal the binding mode of three chemotherapeutic compounds, demonstrate how these molecules open the closed conformation of the transporter, and establish that imatinib is particularly effective in stabilizing the inward facing conformation of ABCG2. Together these studies reveal the previously unrecognized conformational cycle of ABCG2. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:32385283 / PubMed:32385283 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.5 - 4.1 Å |
| Structure data | EMDB-21436, PDB-6vxf: EMDB-21437, PDB-6vxh: EMDB-21438, PDB-6vxi:  EMDB-21439: EMDB-21440, PDB-6vxj:  EMDB-21441: |
| Chemicals |  ChemComp-CLR: 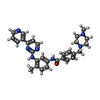 ChemComp-STI: 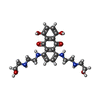 ChemComp-MIX:  ChemComp-RS4: |
| Source |
|
 Keywords Keywords |  TRANSLOCASE / TRANSLOCASE /  ABCG2 / BCRP / ABCG2 / BCRP /  nanodisc / nanodisc /  ABC transporter / TRANSLOCASE/TRANSLOCASE INHIBITOR / ABC transporter / TRANSLOCASE/TRANSLOCASE INHIBITOR /  imatinib / TRANSLOCASE-TRANSLOCASE INHIBITOR complex / imatinib / TRANSLOCASE-TRANSLOCASE INHIBITOR complex /  mitoxantrone / SN38 mitoxantrone / SN38 |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers












