+Search query
-Structure paper
| Title | Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes. |
|---|---|
| Journal, issue, pages | Structure, Vol. 27, Issue 12, Page 1782-11797.e7, Year 2019 |
| Publish date | Dec 3, 2019 |
 Authors Authors | Sanchaita Das / Andrew W Malaby / Agata Nawrotek / Wenhua Zhang / Mahel Zeghouf / Sarah Maslen / Mark Skehel / Srinivas Chakravarthy / Thomas C Irving / Osman Bilsel / Jacqueline Cherfils / David G Lambright /    |
| PubMed Abstract | Membrane dynamic processes require Arf GTPase activation by guanine nucleotide exchange factors (GEFs) with a Sec7 domain. Cytohesin family Arf GEFs function in signaling and cell migration through ...Membrane dynamic processes require Arf GTPase activation by guanine nucleotide exchange factors (GEFs) with a Sec7 domain. Cytohesin family Arf GEFs function in signaling and cell migration through Arf GTPase activation on the plasma membrane and endosomes. In this study, the structural organization of two cytohesins (Grp1 and ARNO) was investigated in solution by size exclusion-small angle X-ray scattering and negative stain-electron microscopy and on membranes by dynamic light scattering, hydrogen-deuterium exchange-mass spectrometry and guanosine diphosphate (GDP)/guanosine triphosphate (GTP) exchange assays. The results suggest that cytohesins form elongated dimers with a central coiled coil and membrane-binding pleckstrin-homology (PH) domains at opposite ends. The dimers display significant conformational heterogeneity, with a preference for compact to intermediate conformations. Phosphoinositide-dependent membrane recruitment is mediated by one PH domain at a time and alters the conformational dynamics to prime allosteric activation by Arf-GTP. A structural model for membrane targeting and allosteric activation of full-length cytohesin dimers is discussed. |
 External links External links |  Structure / Structure /  PubMed:31601460 / PubMed:31601460 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 53.0 Å |
| Structure data | EMDB-20628: Volume 1 for truncated dimeric Cytohesin-3 (Grp1; amino acids 14-399) EMDB-20629, PDB-6u3g: |
| Chemicals | 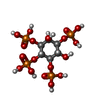 ChemComp-4IP: |
| Source |
|
 Keywords Keywords |  ENDOCYTOSIS / Arf GEF / ENDOCYTOSIS / Arf GEF /  phosphoinositide binding / Sec7 domain / phosphoinositide binding / Sec7 domain /  PH domain PH domain |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers








