+Search query
-Structure paper
| Title | Three-step mechanism of promoter escape by RNA polymerase II. |
|---|---|
| Journal, issue, pages | Mol Cell, Year 2024 |
| Publish date | Apr 5, 2024 |
 Authors Authors | Yumeng Zhan / Frauke Grabbe / Elisa Oberbeckmann / Christian Dienemann / Patrick Cramer /  |
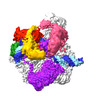
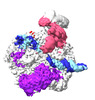
| PubMed Abstract | The transition from transcription initiation to elongation is highly regulated in human cells but remains incompletely understood at the structural level. In particular, it is unclear how ...The transition from transcription initiation to elongation is highly regulated in human cells but remains incompletely understood at the structural level. In particular, it is unclear how interactions between RNA polymerase II (RNA Pol II) and initiation factors are broken to enable promoter escape. Here, we reconstitute RNA Pol II promoter escape in vitro and determine high-resolution structures of initially transcribing complexes containing 8-, 10-, and 12-nt ordered RNAs and two elongation complexes containing 14-nt RNAs. We suggest that promoter escape occurs in three major steps. First, the growing RNA displaces the B-reader element of the initiation factor TFIIB without evicting TFIIB. Second, the rewinding of the transcription bubble coincides with the eviction of TFIIA, TFIIB, and TBP. Third, the binding of DSIF and NELF facilitates TFIIE and TFIIH dissociation, establishing the paused elongation complex. This three-step model for promoter escape fills a gap in our understanding of the initiation-elongation transition of RNA Pol II transcription. |
 External links External links |  Mol Cell / Mol Cell /  PubMed:38604172 PubMed:38604172 |
| Methods | EM (single particle) |
| Resolution | 2.9 - 7.4 Å |
| Structure data | EMDB-19718, PDB-8s51: EMDB-19719, PDB-8s52: EMDB-19720, PDB-8s54:  EMDB-19721: RNA polymerase II initially transcribing complex with an ordered RNA of 8 nt  EMDB-19722: RNA polymerase II initially transcribing complex with an ordered RNA of 10 nt  EMDB-19723: RNA polymerase II initially transcribing complex with an ordered RNA of 12 nt  EMDB-19724: RNA polymerase II early elongation complex bound to TFIIE, TFIIF and TFIIH - state a  EMDB-19725: RNA polymerase II early elongation complex bound to TFIIE, TFIIF and TFIIH - state b EMDB-19726, PDB-8s55: EMDB-19743, PDB-8s5n:  EMDB-19795: RNA polymerase II early elongation complex bound to TFIIE and TFIIF - state a (global map)  EMDB-19796: RNA polymerase II early elongation complex bound to TFIIE and TFIIF - state b (global map)  EMDB-19797: EC14 core Pol II focused map |
| Chemicals |  ChemComp-ZN:  ChemComp-MG: |
| Source |
|
 Keywords Keywords |  TRANSCRIPTION / TRANSCRIPTION /  RNA polymerase II / promoter escape / de novo initiation / de novo transcription RNA polymerase II / promoter escape / de novo initiation / de novo transcription |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers












 unidentified adenovirus
unidentified adenovirus
