+Search query
-Structure paper
| Title | CENP-A and CENP-B collaborate to create an open centromeric chromatin state. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 14, Issue 1, Page 8227, Year 2023 |
| Publish date | Dec 12, 2023 |
 Authors Authors | Harsh Nagpal / Ahmad Ali-Ahmad / Yasuhiro Hirano / Wei Cai / Mario Halic / Tatsuo Fukagawa / Nikolina Sekulić / Beat Fierz /     |
| PubMed Abstract | Centromeres are epigenetically defined via the presence of the histone H3 variant CENP-A. Contacting CENP-A nucleosomes, the constitutive centromere associated network (CCAN) and the kinetochore ...Centromeres are epigenetically defined via the presence of the histone H3 variant CENP-A. Contacting CENP-A nucleosomes, the constitutive centromere associated network (CCAN) and the kinetochore assemble, connecting the centromere to spindle microtubules during cell division. The DNA-binding centromeric protein CENP-B is involved in maintaining centromere stability and, together with CENP-A, shapes the centromeric chromatin state. The nanoscale organization of centromeric chromatin is not well understood. Here, we use single-molecule fluorescence and cryoelectron microscopy (cryoEM) to show that CENP-A incorporation establishes a dynamic and open chromatin state. The increased dynamics of CENP-A chromatin create an opening for CENP-B DNA access. In turn, bound CENP-B further opens the chromatin fiber structure and induces nucleosomal DNA unwrapping. Finally, removal of CENP-A increases CENP-B mobility in cells. Together, our studies show that the two centromere-specific proteins collaborate to reshape chromatin structure, enabling the binding of centromeric factors and establishing a centromeric chromatin state. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:38086807 / PubMed:38086807 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.73 - 6.7 Å |
| Structure data | 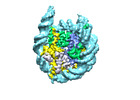 EMDB-18699: Human H3 nucleosome assembled on alpha-satellite DNA (Class1: most wrapped DNA) 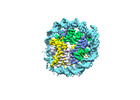 EMDB-18714: Human H3 nucleosome assembled on alpha-satellite DNA (most unwrapped) 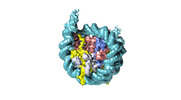 EMDB-18739: Human CENP-A nucleosome assembled on alpha-satellite DNA (most wrapped DNA) 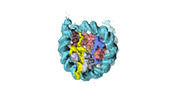 EMDB-18740: Human CENP-A nucleosome assembled on alpha-satellite DNA (partially unwrapped)  EMDB-18745: Human Cenp-A nucleosome assembled on alpha-satellite DNA (most unwrapped DNA) 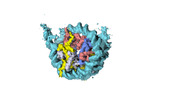 EMDB-18753: Human CENP-A nucleosome assembled on alpha-satellite DNA in complex with CENP-B (most wrapped DNA) 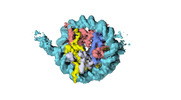 EMDB-18763: Human CENP-A nucleosome assembled on alpha-satellite DNA in complex with CENP-B (partially unwrapped DNA) 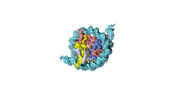 EMDB-18768: Human CENP-A nucleosome assembled on alpha-satellite DNA (most unwrapped DNA) 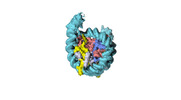 EMDB-18775: Human CENP-A nucleosome assembled on 601 DNA with CENP-B box  EMDB-18776: Human CENP-A nucleosome assembled on 601 DNA with CENP-B box in complex with CENP-B |
| Source |
|
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




 Homo sapiens (human)
Homo sapiens (human)