+Search query
-Structure paper
| Title | ATG9A and ATG2A form a heteromeric complex essential for autophagosome formation. |
|---|---|
| Journal, issue, pages | Mol Cell, Vol. 82, Issue 22, Page 4324-44339.e8, Year 2022 |
| Publish date | Nov 17, 2022 |
 Authors Authors | Alexander R van Vliet / George N Chiduza / Sarah L Maslen / Valerie E Pye / Dhira Joshi / Stefano De Tito / Harold B J Jefferies / Evangelos Christodoulou / Chloë Roustan / Emma Punch / Javier H Hervás / Nicola O'Reilly / J Mark Skehel / Peter Cherepanov / Sharon A Tooze /   |
| PubMed Abstract | ATG9A and ATG2A are essential core members of the autophagy machinery. ATG9A is a lipid scramblase that allows equilibration of lipids across a membrane bilayer, whereas ATG2A facilitates lipid flow ...ATG9A and ATG2A are essential core members of the autophagy machinery. ATG9A is a lipid scramblase that allows equilibration of lipids across a membrane bilayer, whereas ATG2A facilitates lipid flow between tethered membranes. Although both have been functionally linked during the formation of autophagosomes, the molecular details and consequences of their interaction remain unclear. By combining data from peptide arrays, crosslinking, and hydrogen-deuterium exchange mass spectrometry together with cryoelectron microscopy, we propose a molecular model of the ATG9A-2A complex. Using this integrative structure modeling approach, we identify several interfaces mediating ATG9A-2A interaction that would allow a direct transfer of lipids from ATG2A into the lipid-binding perpendicular branch of ATG9A. Mutational analyses combined with functional activity assays demonstrate their importance for autophagy, thereby shedding light on this protein complex at the heart of autophagy. |
 External links External links |  Mol Cell / Mol Cell /  PubMed:36347259 PubMed:36347259 |
| Methods | EM (single particle) |
| Resolution | 14.7 - 32.89 Å |
| Structure data | 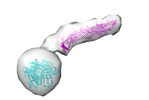 EMDB-15604: ATG9A and ATG2A form a heteromeric complex essential for autophagosome formation 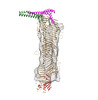 EMDB-15605: Low resolution 3D reconstruction of ATG2A from cryo-EM |
| Source |
|
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




 Homo sapiens (human)
Homo sapiens (human)