+Search query
-Structure paper
| Title | Visualizing translation dynamics at atomic detail inside a bacterial cell. |
|---|---|
| Journal, issue, pages | Nature, Vol. 610, Issue 7930, Page 205-211, Year 2022 |
| Publish date | Sep 28, 2022 |
 Authors Authors | Liang Xue / Swantje Lenz / Maria Zimmermann-Kogadeeva / Dimitry Tegunov / Patrick Cramer / Peer Bork / Juri Rappsilber / Julia Mahamid /    |
| PubMed Abstract | Translation is the fundamental process of protein synthesis and is catalysed by the ribosome in all living cells. Here we use advances in cryo-electron tomography and sub-tomogram analysis to ...Translation is the fundamental process of protein synthesis and is catalysed by the ribosome in all living cells. Here we use advances in cryo-electron tomography and sub-tomogram analysis to visualize the structural dynamics of translation inside the bacterium Mycoplasma pneumoniae. To interpret the functional states in detail, we first obtain a high-resolution in-cell average map of all translating ribosomes and build an atomic model for the M. pneumoniae ribosome that reveals distinct extensions of ribosomal proteins. Classification then resolves 13 ribosome states that differ in their conformation and composition. These recapitulate major states that were previously resolved in vitro, and reflect intermediates during active translation. On the basis of these states, we animate translation elongation inside native cells and show how antibiotics reshape the cellular translation landscapes. During translation elongation, ribosomes often assemble in defined three-dimensional arrangements to form polysomes. By mapping the intracellular organization of translating ribosomes, we show that their association into polysomes involves a local coordination mechanism that is mediated by the ribosomal protein L9. We propose that an extended conformation of L9 within polysomes mitigates collisions to facilitate translation fidelity. Our work thus demonstrates the feasibility of visualizing molecular processes at atomic detail inside cells. |
 External links External links |  Nature / Nature /  PubMed:36171285 / PubMed:36171285 /  PubMed Central PubMed Central |
| Methods | EM (subtomogram averaging) |
| Resolution | 3.4 - 23.0 Å |
| Structure data | EMDB-13234, PDB-7p6z: EMDB-13272, PDB-7pah: EMDB-13273, PDB-7pai: EMDB-13274, PDB-7paj: EMDB-13275, PDB-7pak: EMDB-13276, PDB-7pal: EMDB-13277, PDB-7pam: EMDB-13278, PDB-7pan: EMDB-13279, PDB-7pao: EMDB-13280, PDB-7paq: EMDB-13281, PDB-7par: EMDB-13282, PDB-7pas:  EMDB-13283: 70S ribosome class with unexplained density near P site in Mycoplasma pneumoniae cells  EMDB-13284: 70S ribosome with dim 30S subunit in Mycoplasma pneumoniae cells EMDB-13285, PDB-7pat: EMDB-13286, PDB-7pau:  EMDB-13287: polysome, ribosome pair (conformation 1) in Mycoplasma pneumoniae cells  EMDB-13288: polysome, ribosome pair (conformation 2) in Mycoplasma pneumoniae cells  EMDB-13289: polysome, ribosome pair (conformation 3) in Mycoplasma pneumoniae cells EMDB-13410, PDB-7ph9: EMDB-13411, PDB-7pha: EMDB-13412, PDB-7phb: EMDB-13413, PDB-7phc:  EMDB-13414: 70S ribosome with dim 30S subunit in chloramphenicol-treated Mycoplasma pneumoniae cells  EMDB-13431: free 50S in spectinomycin-treated Mycoplasma pneumoniae cells EMDB-13432, PDB-7pi8: EMDB-13433, PDB-7pi9: EMDB-13434, PDB-7pia: EMDB-13435, PDB-7pib: EMDB-13436, PDB-7pic: EMDB-13445, PDB-7pio: EMDB-13446, PDB-7pip: EMDB-13447, PDB-7piq: EMDB-13448, PDB-7pir: EMDB-13449, PDB-7pis: EMDB-13450, PDB-7pit:  EMDB-13451: 70S ribosome with dim 30S subunit in pseudouridimycin-treated Mycoplasma pneumoniae cells  EMDB-13452: free 50S in pseudouridimycin-treated Mycoplasma pneumoniae cells  PDB-7ooc:  PDB-7ood: |
| Chemicals |  ChemComp-ZN:  ChemComp-K:  ChemComp-MG: 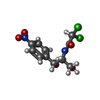 ChemComp-CLM: 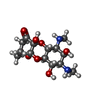 ChemComp-SCM: |
| Source |
|
 Keywords Keywords |  RIBOSOME / In-cell cryo-electron tomography chloramphenicol-treated sub-tomogram analysis / In-cell cryo-electron tomography / sub-tomogram analysis / RIBOSOME / In-cell cryo-electron tomography chloramphenicol-treated sub-tomogram analysis / In-cell cryo-electron tomography / sub-tomogram analysis /  TRANSLATION / translation intermediate state / translation termination / TRANSLATION / translation intermediate state / translation termination /  chloramphenicol / chloramphenicol /  spectinomycin / pseudouridimycin spectinomycin / pseudouridimycin |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers





























































 mycoplasma pneumoniae (strain atcc 29342 / m129) (bacteria)
mycoplasma pneumoniae (strain atcc 29342 / m129) (bacteria)