+Search query
-Structure paper
| Title | Recognition of methamphetamine and other amines by trace amine receptor TAAR1. |
|---|---|
| Journal, issue, pages | Nature, Vol. 624, Issue 7992, Page 663-671, Year 2023 |
| Publish date | Nov 7, 2023 |
 Authors Authors | Heng Liu / You Zheng / Yue Wang / Yumeng Wang / Xinheng He / Peiyu Xu / Sijie Huang / Qingning Yuan / Xinyue Zhang / Ling Wang / Kexin Jiang / Hong Chen / Zhen Li / Wenbin Liu / Sheng Wang / H Eric Xu / Fei Xu /  |
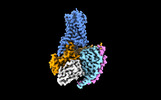
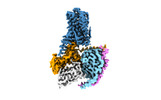
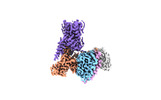
| PubMed Abstract | Trace amine-associated receptor 1 (TAAR1), the founding member of a nine-member family of trace amine receptors, is responsible for recognizing a range of biogenic amines in the brain, including the ...Trace amine-associated receptor 1 (TAAR1), the founding member of a nine-member family of trace amine receptors, is responsible for recognizing a range of biogenic amines in the brain, including the endogenous β-phenylethylamine (β-PEA) as well as methamphetamine, an abused substance that has posed a severe threat to human health and society. Given its unique physiological role in the brain, TAAR1 is also an emerging target for a range of neurological disorders including schizophrenia, depression and drug addiction. Here we report structures of human TAAR1-G-protein complexes bound to methamphetamine and β-PEA as well as complexes bound to RO5256390, a TAAR1-selective agonist, and SEP-363856, a clinical-stage dual agonist for TAAR1 and serotonin receptor 5-HTR (refs. ). Together with systematic mutagenesis and functional studies, the structures reveal the molecular basis of methamphetamine recognition and underlying mechanisms of ligand selectivity and polypharmacology between TAAR1 and other monoamine receptors. We identify a lid-like extracellular loop 2 helix/loop structure and a hydrogen-bonding network in the ligand-binding pockets, which may contribute to the ligand recognition in TAAR1. These findings shed light on the ligand recognition mode and activation mechanism for TAAR1 and should guide the development of next-generation therapeutics for drug addiction and various neurological disorders. |
 External links External links |  Nature / Nature /  PubMed:37935377 PubMed:37935377 |
| Methods | EM (single particle) |
| Resolution | 2.6 - 3.0 Å |
| Structure data | EMDB-37347, PDB-8w87: EMDB-37348, PDB-8w88: EMDB-37349, PDB-8w89: EMDB-37350, PDB-8w8a: EMDB-37351, PDB-8w8b: |
| Chemicals | 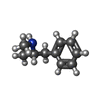 ChemComp-B40:  ChemComp-CLR:  ChemComp-UJL:  ChemComp-PEA:  ChemComp-HOH:  ChemComp-T5U:  ChemComp-PLM: |
| Source |
|
 Keywords Keywords |  MEMBRANE PROTEIN / MEMBRANE PROTEIN /  TAAR1 / TAAR1 /  METH / METH /  GPCR / SEP-363856 / antipsychotics / GPCR / SEP-363856 / antipsychotics /  PEA / R05256390 / PEA / R05256390 /  5-HT1A / SEP363856 5-HT1A / SEP363856 |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers