+Search query
-Structure paper
| Title | Structural mechanism of LIN28B nucleosome targeting by OCT4. |
|---|---|
| Journal, issue, pages | Mol Cell, Vol. 83, Issue 12, Page 1970-11982.e6, Year 2023 |
| Publish date | Jun 15, 2023 |
 Authors Authors | Ruifang Guan / Tengfei Lian / Bing-Rui Zhou / David Wheeler / Yawen Bai /  |
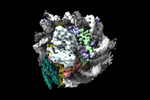
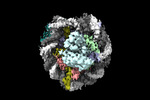
| PubMed Abstract | Pioneer transcription factors are essential for cell fate changes by targeting closed chromatin. OCT4 is a crucial pioneer factor that can induce cell reprogramming. However, the structural basis of ...Pioneer transcription factors are essential for cell fate changes by targeting closed chromatin. OCT4 is a crucial pioneer factor that can induce cell reprogramming. However, the structural basis of how pioneer factors recognize the in vivo nucleosomal DNA targets is unknown. Here, we determine the high-resolution structures of the nucleosome containing human LIN28B DNA and its complexes with the OCT4 DNA binding region. Three OCT4s bind the pre-positioned nucleosome by recognizing non-canonical DNA sequences. Two use their POUS domains while the other uses the POUS-loop-POUHD region; POUHD serves as a wedge to unwrap ∼25 base pair DNA. Our analysis of previous genomic data and determination of the ESRRB-nucleosome-OCT4 structure confirmed the generality of these structural features. Moreover, biochemical studies suggest that multiple OCT4s cooperatively open the H1-condensed nucleosome array containing the LIN28B nucleosome. Thus, our study suggests a mechanism of how OCT4 can target the nucleosome and open closed chromatin. |
 External links External links |  Mol Cell / Mol Cell /  PubMed:37327775 / PubMed:37327775 /  PubMed Central PubMed Central |
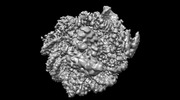
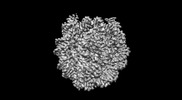
| Methods | EM (single particle) |
| Resolution | 2.6 - 7.9 Å |
| Structure data | EMDB-26258, PDB-7u0g: EMDB-26260, PDB-7u0i: EMDB-26261, PDB-7u0j: EMDB-27483, PDB-8dk5: EMDB-40683, PDB-8sps:  EMDB-40684: Low resolution reconstruction of ESRRB nucleosome bound OCT4 at site a and site b EMDB-40686, PDB-8spu:  EMDB-40691: Focused classification of site c OCT4 |
| Source |
|
 Keywords Keywords | TRANSCRIPTION/DNA /  nucleosome / nucleosome /  transcription factor / transcription factor /  transcription / CHROMATIN BINDING PROTEIN-DNA complex / TRANSCRIPTION-DNA complex / transcription / CHROMATIN BINDING PROTEIN-DNA complex / TRANSCRIPTION-DNA complex /  GENE REGULATION GENE REGULATION |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers