+Search query
-Structure paper
| Title | Structural analysis reveals TLR7 dynamics underlying antagonism. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 11, Issue 1, Page 5204, Year 2020 |
| Publish date | Oct 15, 2020 |
 Authors Authors | Shingo Tojo / Zhikuan Zhang / Hiroyuki Matsui / Masahiro Tahara / Mitsunori Ikeguchi / Mami Kochi / Mami Kamada / Hideki Shigematsu / Akihisa Tsutsumi / Naruhiko Adachi / Takuma Shibata / Masaki Yamamoto / Masahide Kikkawa / Toshiya Senda / Yoshiaki Isobe / Umeharu Ohto / Toshiyuki Shimizu /  |
| PubMed Abstract | Toll-like receptor 7 (TLR7) recognizes both microbial and endogenous RNAs and nucleosides. Aberrant activation of TLR7 has been implicated in several autoimmune diseases including systemic lupus ...Toll-like receptor 7 (TLR7) recognizes both microbial and endogenous RNAs and nucleosides. Aberrant activation of TLR7 has been implicated in several autoimmune diseases including systemic lupus erythematosus (SLE). Here, by modifying potent TLR7 agonists, we develop a series of TLR7-specific antagonists as promising therapeutic agents for SLE. These compounds protect mice against lethal autoimmunity. Combining crystallography and cryo-electron microscopy, we identify the open conformation of the receptor and reveal the structural equilibrium between open and closed conformations that underlies TLR7 antagonism, as well as the detailed mechanism by which TLR7-specific antagonists bind to their binding pocket in TLR7. Our work provides small-molecule TLR7-specific antagonists and suggests the TLR7-targeting strategy for treating autoimmune diseases. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:33060576 / PubMed:33060576 /  PubMed Central PubMed Central |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 2.6 - 4.4 Å |
| Structure data |  EMDB-0999:  EMDB-1000:  EMDB-30000:  EMDB-30001: EMDB-30002, PDB-6lw1:  PDB-6lvx:  PDB-6lvy:  PDB-6lvz:  PDB-6lw0: |
| Chemicals |  ChemComp-EWL:  ChemComp-NAG:  ChemComp-SO4:  ChemComp-HOH:  ChemComp-EWU:  ChemComp-EWX: 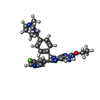 ChemComp-EX0: 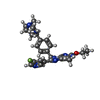 ChemComp-EX3: |
| Source |
|
 Keywords Keywords | IMMUNE SYSTEM / TLR7 / agonist / antagonist |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers






