+Search query
-Structure paper
| Title | Mapping Neutralizing Antibody Epitope Specificities to an HIV Env Trimer in Immunized and in Infected Rhesus Macaques. |
|---|---|
| Journal, issue, pages | Cell Rep, Vol. 32, Issue 10, Page 108122, Year 2020 |
| Publish date | Sep 8, 2020 |
 Authors Authors | Fangzhu Zhao / Collin Joyce / Alison Burns / Bartek Nogal / Christopher A Cottrell / Alejandra Ramos / Trevor Biddle / Matthias Pauthner / Rebecca Nedellec / Huma Qureshi / Rosemarie Mason / Elise Landais / Bryan Briney / Andrew B Ward / Dennis R Burton / Devin Sok /  |
| PubMed Abstract | BG505 SOSIP is a well-characterized near-native recombinant HIV Envelope (Env) trimer that holds promise as part of a sequential HIV immunogen regimen to induce broadly neutralizing antibodies (bnAbs) ...BG505 SOSIP is a well-characterized near-native recombinant HIV Envelope (Env) trimer that holds promise as part of a sequential HIV immunogen regimen to induce broadly neutralizing antibodies (bnAbs). Rhesus macaques are considered the most appropriate pre-clinical animal model for monitoring antibody (Ab) responses. Accordingly, we report here the isolation of 45 BG505 autologous neutralizing antibodies (nAbs) with multiple specificities from SOSIP-immunized and BG505 SHIV-infected rhesus macaques. We associate the most potent neutralization with two epitopes: the C3/V5 and V1/V3 regions. We show that all of the nAbs bind in close proximity to known bnAb epitopes and might therefore sterically hinder elicitation of bnAbs. We also identify a "public clonotype" that targets the immunodominant C3/V5 nAb epitope, which suggests that common antibody rearrangements might help determine humoral responses to Env immunogens. The results highlight important considerations for vaccine design in anticipation of results of the BG505 SOSIP trimer in clinical trials. |
 External links External links |  Cell Rep / Cell Rep /  PubMed:32905766 / PubMed:32905766 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 17.4 - 27.1 Å |
| Structure data |  EMDB-22372: 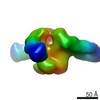 EMDB-22373: 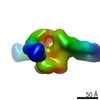 EMDB-22374:  EMDB-22385: 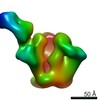 EMDB-22387: 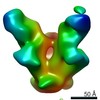 EMDB-22388: |
| Source |
|
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers





 Human immunodeficiency virus 1
Human immunodeficiency virus 1
