+Search query
-Structure paper
| Title | Characterization of virus-like particles in GARDASIL® by cryo transmission electron microscopy. |
|---|---|
| Journal, issue, pages | Hum Vaccin Immunother, Vol. 10, Issue 3, Page 734-739, Year 2014 |
| Publish date | Dec 3, 2013 |
 Authors Authors | Qinjian Zhao / Clinton S Potter / Bridget Carragher / Gabriel Lander / Jaime Sworen / Victoria Towne / Dicky Abraham / Paul Duncan / Michael W Washabaugh / Robert D Sitrin /  |
| PubMed Abstract | Cryo-transmission electron microscopy (cryoTEM) is a powerful characterization method for assessing the structural properties of biopharmaceutical nanoparticles, including Virus Like Particle-based ...Cryo-transmission electron microscopy (cryoTEM) is a powerful characterization method for assessing the structural properties of biopharmaceutical nanoparticles, including Virus Like Particle-based vaccines. We demonstrate the method using the Human Papilloma Virus (HPV) VLPs in GARDASIL®. CryoTEM, coupled to automated data collection and analysis, was used to acquire images of the particles in their hydrated state, determine their morphological characteristics, and confirm the integrity of the particles when absorbed to aluminum adjuvant. In addition, we determined the three-dimensional structure of the VLPs, both alone and when interacting with neutralizing antibodies. Two modes of binding of two different neutralizing antibodies were apparent; for HPV type 11 saturated with H11.B2, 72 potential Fab binding sites were observed at the center of each capsomer, whereas for HPV 16 interacting with H16.V5, it appears that 60 pentamers (each neighboring 6 other pentamers) bind five Fabs per pentamer, for the total of 300 potential Fab binding sites per VLP. |
 External links External links |  Hum Vaccin Immunother / Hum Vaccin Immunother /  PubMed:24299977 / PubMed:24299977 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 13.0 - 20.0 Å |
| Structure data |  EMDB-5837: 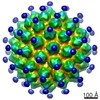 EMDB-5838:  EMDB-5839:  EMDB-5840: |
| Source |
|
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



