+Search query
-Structure paper
| Title | Reprogramming an ATP-driven protein machine into a light-gated nanocage. |
|---|---|
| Journal, issue, pages | Nat Nanotechnol, Vol. 8, Issue 12, Page 928-932, Year 2013 |
| Publish date | Nov 24, 2013 |
 Authors Authors | Daniel Hoersch / Soung-Hun Roh / Wah Chiu / Tanja Kortemme /  |
| PubMed Abstract | Natural protein assemblies have many sophisticated architectures and functions, creating nanoscale storage containers, motors and pumps. Inspired by these systems, protein monomers have been ...Natural protein assemblies have many sophisticated architectures and functions, creating nanoscale storage containers, motors and pumps. Inspired by these systems, protein monomers have been engineered to self-assemble into supramolecular architectures including symmetrical, metal-templated and cage-like structures. The complexity of protein machines, however, has made it difficult to create assemblies with both defined structures and controllable functions. Here we report protein assemblies that have been engineered to function as light-controlled nanocontainers. We show that an adenosine-5'-triphosphate-driven group II chaperonin, which resembles a barrel with a built-in lid, can be reprogrammed to open and close on illumination with different wavelengths of light. By engineering photoswitchable azobenzene-based molecules into the structure, light-triggered changes in interatomic distances in the azobenzene moiety are able to drive large-scale conformational changes of the protein assembly. The different states of the assembly can be visualized with single-particle cryo-electron microscopy, and the nanocages can be used to capture and release non-native cargos. Similar strategies that switch atomic distances with light could be used to build other controllable nanoscale machines. |
 External links External links |  Nat Nanotechnol / Nat Nanotechnol /  PubMed:24270642 / PubMed:24270642 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 15.6 - 18.9 Å |
| Structure data |  EMDB-5767: 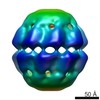 EMDB-5768: 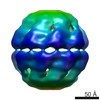 EMDB-5769:  EMDB-5770: |
| Source |
|
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




 Methanococcus maripaludis (archaea)
Methanococcus maripaludis (archaea)