+Search query
-Structure paper
| Title | Characterization of the insertase for β-barrel proteins of the outer mitochondrial membrane. |
|---|---|
| Journal, issue, pages | J Cell Biol, Vol. 199, Issue 4, Page 599-611, Year 2012 |
| Publish date | Nov 12, 2012 |
 Authors Authors | Astrid Klein / Lars Israel / Sebastian W K Lackey / Frank E Nargang / Axel Imhof / Wolfgang Baumeister / Walter Neupert / Dennis R Thomas /  |
| PubMed Abstract | The TOB-SAM complex is an essential component of the mitochondrial outer membrane that mediates the insertion of β-barrel precursor proteins into the membrane. We report here its isolation and ...The TOB-SAM complex is an essential component of the mitochondrial outer membrane that mediates the insertion of β-barrel precursor proteins into the membrane. We report here its isolation and determine its size, composition, and structural organization. The complex from Neurospora crassa was composed of Tob55-Sam50, Tob38-Sam35, and Tob37-Sam37 in a stoichiometry of 1:1:1 and had a molecular mass of 140 kD. A very minor fraction of the purified complex was associated with one Mdm10 protein. Using molecular homology modeling for Tob55 and cryoelectron microscopy reconstructions of the TOB complex, we present a model of the TOB-SAM complex that integrates biochemical and structural data. We discuss our results and the structural model in the context of a possible mechanism of the TOB insertase. |
 External links External links |  J Cell Biol / J Cell Biol /  PubMed:23128244 / PubMed:23128244 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 13.4 - 15.6 Å |
| Structure data |  EMDB-2195: 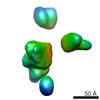 EMDB-2196:  EMDB-2197:  EMDB-2200:  EMDB-2201:  EMDB-2202:  EMDB-2203: |
| Source |
|
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




 Neurospora crassa (fungus)
Neurospora crassa (fungus)