+Search query
-Structure paper
| Title | Structural insights on the plant salt-overly-sensitive 1 (SOS1) Na(+)/H(+) antiporter. |
|---|---|
| Journal, issue, pages | J Mol Biol, Vol. 424, Issue 5, Page 283-294, Year 2012 |
| Publish date | Dec 14, 2012 |
 Authors Authors | Rafael Núñez-Ramírez / María José Sánchez-Barrena / Irene Villalta / Juan F Vega / Jose M Pardo / Francisco J Quintero / Javier Martinez-Salazar / Armando Albert /  |
| PubMed Abstract | The Arabidopsisthaliana Na(+)/H(+) antiporter salt-overly-sensitive 1 (SOS1) is essential to maintain low intracellular levels of toxic Na(+) under salt stress. Available data show that the plant ...The Arabidopsisthaliana Na(+)/H(+) antiporter salt-overly-sensitive 1 (SOS1) is essential to maintain low intracellular levels of toxic Na(+) under salt stress. Available data show that the plant SOS2 protein kinase and its interacting activator, the SOS3 calcium-binding protein, function together in decoding calcium signals elicited by salt stress and regulating the phosphorylation state and the activity of SOS1. Molecular genetic studies have shown that the activation implies a domain reorganization of the antiporter cytosolic moiety, indicating that there is a clear relationship between function and molecular structure of the antiporter. To provide information on this issue, we have carried out in vivo and in vitro studies on the oligomerization state of SOS1. In addition, we have performed electron microscopy and single-particle reconstruction of negatively stained full-length and active SOS1. Our studies show that the protein is a homodimer that contains a membrane domain similar to that found in other antiporters of the family and an elongated, large, and structured cytosolic domain. Both the transmembrane (TM) and cytosolic moieties contribute to the dimerization of the antiporter. The close contacts between the TM and the cytosolic domains provide a link between regulation and transport activity of the antiporter. |
 External links External links |  J Mol Biol / J Mol Biol /  PubMed:23022605 PubMed:23022605 |
| Methods | EM (single particle) |
| Resolution | 25.9 Å |
| Structure data | 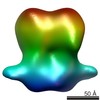 EMDB-2181:  EMDB-2182: |
| Source |
|
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




 Arabidopsis thaliana (thale cress)
Arabidopsis thaliana (thale cress)