+Search query
-Structure paper
| Title | Proteome organization in a genome-reduced bacterium. |
|---|---|
| Journal, issue, pages | Science, Vol. 326, Issue 5957, Page 1235-1240, Year 2009 |
| Publish date | Nov 27, 2009 |
 Authors Authors | Sebastian Kühner / Vera van Noort / Matthew J Betts / Alejandra Leo-Macias / Claire Batisse / Michaela Rode / Takuji Yamada / Tobias Maier / Samuel Bader / Pedro Beltran-Alvarez / Daniel Castaño-Diez / Wei-Hua Chen / Damien Devos / Marc Güell / Tomas Norambuena / Ines Racke / Vladimir Rybin / Alexander Schmidt / Eva Yus / Ruedi Aebersold / Richard Herrmann / Bettina Böttcher / Achilleas S Frangakis / Robert B Russell / Luis Serrano / Peer Bork / Anne-Claude Gavin /  |
| PubMed Abstract | The genome of Mycoplasma pneumoniae is among the smallest found in self-replicating organisms. To study the basic principles of bacterial proteome organization, we used tandem affinity purification- ...The genome of Mycoplasma pneumoniae is among the smallest found in self-replicating organisms. To study the basic principles of bacterial proteome organization, we used tandem affinity purification-mass spectrometry (TAP-MS) in a proteome-wide screen. The analysis revealed 62 homomultimeric and 116 heteromultimeric soluble protein complexes, of which the majority are novel. About a third of the heteromultimeric complexes show higher levels of proteome organization, including assembly into larger, multiprotein complex entities, suggesting sequential steps in biological processes, and extensive sharing of components, implying protein multifunctionality. Incorporation of structural models for 484 proteins, single-particle electron microscopy, and cellular electron tomograms provided supporting structural details for this proteome organization. The data set provides a blueprint of the minimal cellular machinery required for life. |
 External links External links |  Science / Science /  PubMed:19965468 PubMed:19965468 |
| Methods | EM (single particle) |
| Resolution | 21.5 - 25.0 Å |
| Structure data | 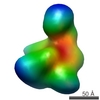 EMDB-1637:  EMDB-1638: 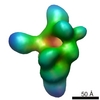 EMDB-1639: |
| Source |
|
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



 Mycoplasma pneumoniae (Filterable agent of primary atypical pneumonia)
Mycoplasma pneumoniae (Filterable agent of primary atypical pneumonia)